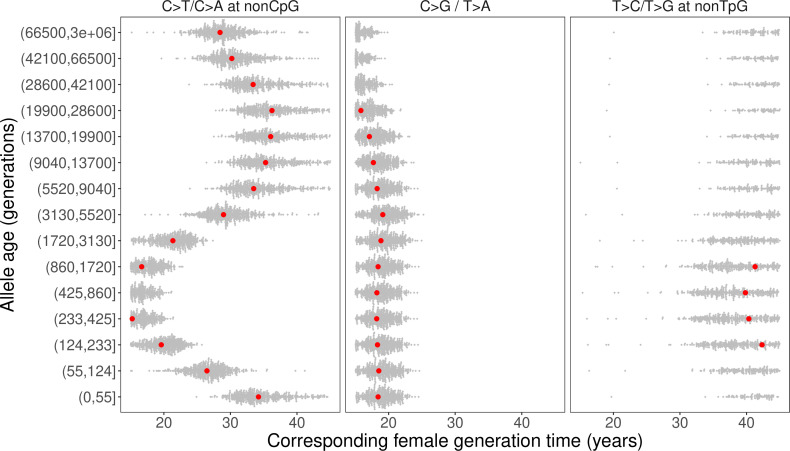
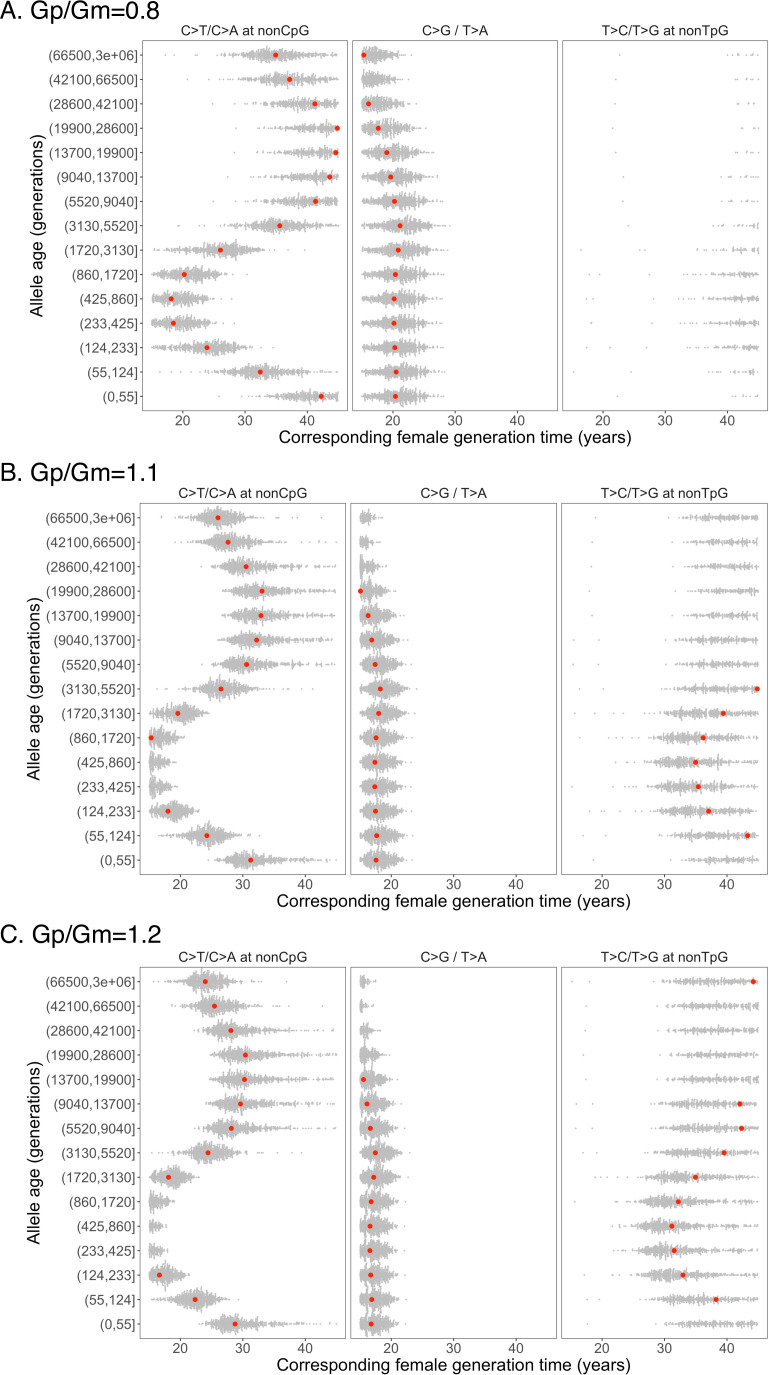
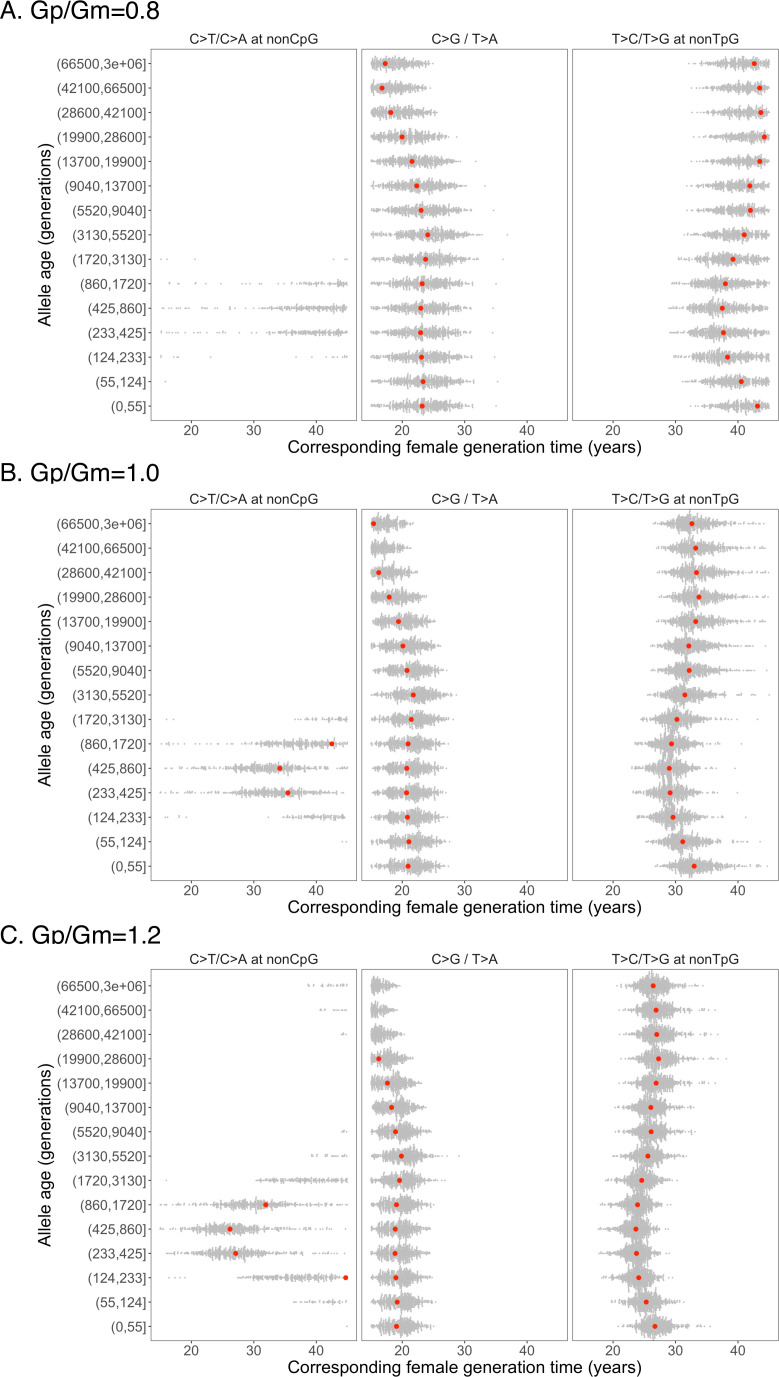
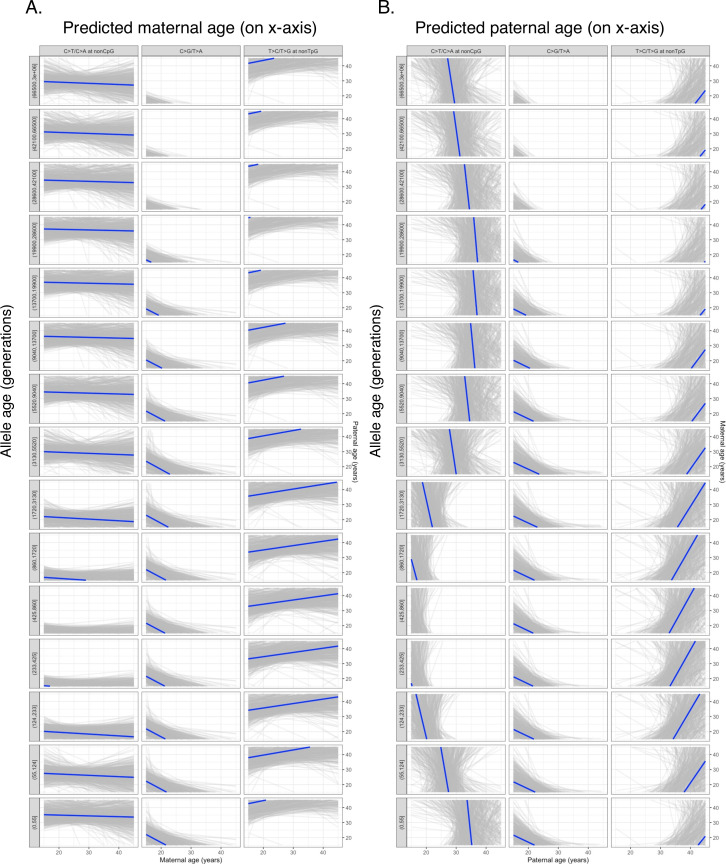
Under the assumption of linear parental age effects on the mutation rate of each mutation type, a specific value of pairwise polymorphism ratio places a linear constraint on the values of paternal and maternal reproductive ages (denoted by Gp and Gm), which is represented by a line in each plot. Blue lines represent predicted constraints based on the maximum likelihood estimators of mutation parameters estimated from de novo mutation (DNM) data (
Halldorsson et al., 2019); gray lines show constraints from 500 bootstrap replicates by resampling trios with replacement. Panels (
A) and (
B) show the same results but with Gm and Gp shown on the x- and y-axes, respectively, in order to illustrate their temporal trends. Consistent with results shown in
Figure 3—figure supplement 3, the nonTpG T>C/T>G and non-CpG C>T/C>A ratios are relatively insensitive to Gm and largely determined by Gp, so the temporal trends of these two mutation ratios in panel (
B) mirror those of
Figure 4. The C>G/T>A depends on both Gp and Gm, so its temporal trend in both panels (
A) and (
B) mirrors that shown in
Figure 4. Note that for each time window the gray areas predicted by three polymorphism ratios barely, if at all, overlap, suggesting that no combination of (Gp, Gm) values can explain the three observed polymorphism ratios simultaneously. In addition, the temporal trends predicted by the three polymorphism ratios disagree with each other.