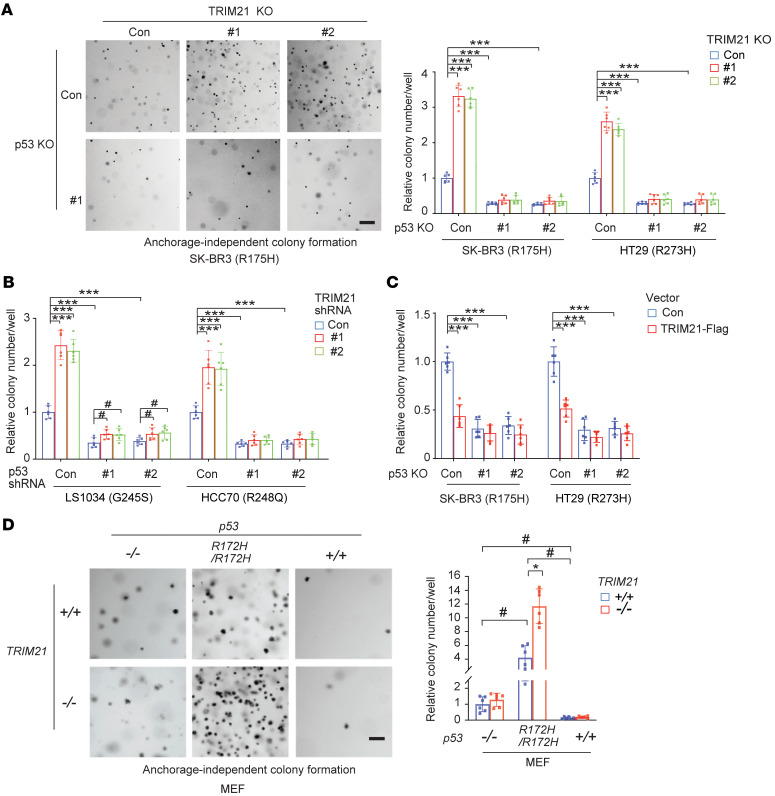
Figure 4. TRIM21 inhibits mutp53 GOF in promoting anchorage-independent growth of cancer cells.
(A) TRIM21 KO enhanced anchorage-independent growth of SK-BR3 and HT29 cells, which was largely abolished by mutp53 KO. The control, TRIM21 KO, mutp53 KO, and TRIM21 + mutp53 double-KO cells were employed for anchorage-independent growth assays in soft agar. Left: Representative images of anchorage-independent growth of SK-BR3 cells. Right: Summary of relative cell colony numbers in soft agar. (B) Knockdown of endogenous TRIM21 by shRNA vectors enhanced anchorage-independent growth of LS1034 and HCC70 cells, which was largely abolished by mutp53 knockdown. The control, TRIM21 knockdown, mutp53 knockdown, and TRIM21 + mutp53 double-knockdown cells were employed for assays. (C) Ectopic TRIM21-Flag expression inhibited the anchorage-independent growth of SK-BR3 and HT29 cells, which was largely abolished by mutp53 KO. Control and mutp53 KO cells were transduced with control or TRIM21-Flag expression vectors for assays. (D) TRIM21 loss enhanced mutp53 GOF activity in promoting anchorage-independent growth of E1A/RasV12-transformed MEFs. The E1A/RasV12-transformed p53R172H/R172H, p53−/−, and p53+/+ MEFs with or without TRIM21 loss (TRIM21−/− or TRIM21+/+) were employed for assays. Left: Representative images of anchorage-independent growth of MEFs. Data are shown as the mean ± SD (n = 6). ANOVA followed by Dunnett’s test. #P < 0.05; *P < 0.01; ***P < 0.0001. Scale bars: 200 μm (A and D).