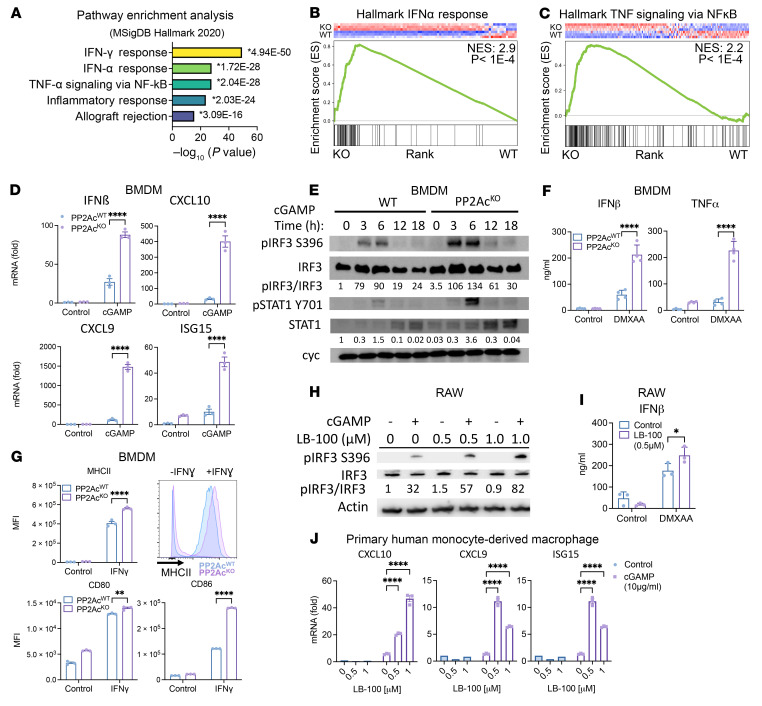
Figure 1. PP2A negatively regulates STING-Type I IFN signaling pathway.
(A) Pathway enrichment analysis of RNA-Seq of PP2AcKO and PP2AcWT BMDM treated with cGAMP (10 μg/mL) for 4 hours (n = 3 per group) showing the top 5 enriched pathways ranked with highest –log10 P value using differentially upregulated genes in PP2AcKO compared with PP2AcWT BMDM (Log2 fold change (log2FC) > 1, FDR < 0.01). * indicates the P value for each individual pathway. (B and C) GSEA plots for Type I IFN (B) and TNF (C) signatures between cGAMP-treated PP2AcKO versus PP2AcWT BMDM. (D) BMDM were harvested 4 hours after cGAMP stimulation (10 μg/mL). Expression of IFNβ and IFN response genes (CXCL10, CXCL9, and ISG15) were measured via reverse transcription PCR. (E) Protein expression of BMDM was analyzed by immunoblotting after cGAMP (10 μg/mL) treatment. (F) PP2AcKO and PP2AcWT BMDM were stimulated with DMXAA (10 μg/mL) for 48 hours, cytokine concentrations were measured in culture supernatant. (G) PP2AcKO and PP2AcWT BMDM were treated with IFNγ (10 ng/mL) for 24 hours, expressions of CD80, CD86, and MHCII were measured by FACS. Representative FACS plot of MHCII expression ± IFNγ treatment. (H) RAW cells were pretreated with the PP2A inhibitor LB-100 for 2 hours before stimulated with cGAMP (10 μg/mL) for 4 hours. Protein expression was analyzed by immunoblotting. (I) RAW cells were pretreated with LB-100 for 2 hours before stimulated with DMXAA (10 μg/mL) for 48 hours. Cytokine concentrations were measured in culture supernatant. (J) PBMCs were treated with M-CSF (50 ng/mL) for 6 days to derive macrophages. Cells were then pretreated with LB-100 at the indicated dosage for 1.5 hours prior to cGAMP (10 μg/mL) treatment. Expression of IFN response genes (CXCL10, CXCL9, and ISG15) were measured via real time PCR. Data are from 1 experiment representative of at least 2 (B–I) and 1 (J) independent experiments with similar results. Error bars depict SEM. P values were calculated by unpaired 2-tailed t test *P < 0.05,**P < 0.01, ***P < 0.001, ****P < 0.0001.