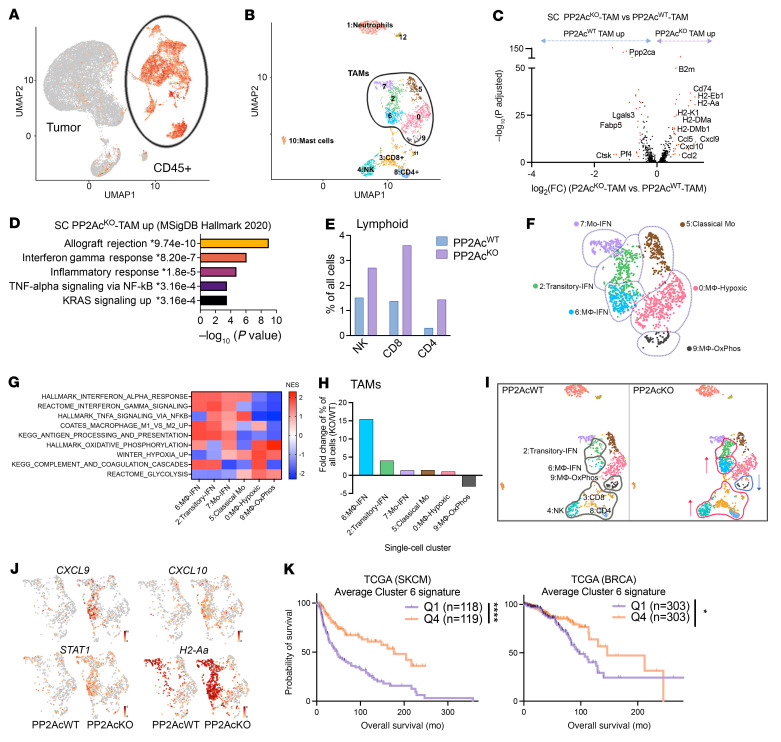
Figure 4. scRNA-Seq of s.c. SB28 tumor.
LysMcrePP2Acfl/fl or WT mice were inoculated with SB28 tumor subcutaneously (0.1 × 106 cells) or orthotopically in the brain (3 × 104 cells). On day 18, 3 tumors per group were pooled and analyzed by scRNA-Seq. (A) UMAP analyses were performed on 26,023 cells from all 4 groups. (B) UMAP of CD45+ immune cells of s.c tumors. Canonical markers were used to identify major immune populations. (C) Volcano plots showing DEGs (−log10 (adjusted P) > 2, log2FC > 0.5) in CD68+TAMs between tumors from LysMcrePP2Acfl/fl or WT mice. P values were adjusted using Bonferroni’s correction. Upregulated genes related to antigen presentation and IFN signaling are labelled. (D) Pathway enrichment analysis performed using Enrichr on upregulated DEGs in tumors from LysMcrePP2Acfl/fl mice. Top 5 enriched biological processes ranked by –log(P). (E) Percentage of lymphoid (CD4, CD8, and NK) cells of all cells. (F) Overview of TAM subsets with 6 subclusters: subcluster 0, hypoxic macrophage; subcluster 2, transitory-IFN; subcluster 5, classical monocyte; subcluster 6, IFN macrophage; subcluster 7, IFN monocytes; and subcluster 9, oxidative phosphorylation (Ox-Phos) macrophage. (G) Heatmap of Normalized Enrichment Score (NES) from GSEA of TAMs cluster. (H) Fold change in frequency of the 6 TAMs clusters. (I) UMAP of immune cells highlighting the clusters that are altered between tumors from LysMcrePP2Acfl/fl or WT mice. (J) UMAP of TAMS highlight IFN-response genes (CXCL9, CXCL10, STAT1, and H2-Aa) in TAMs from LysMcrePP2Acfl/fl or WT mice. (K) Average expression level of cluster 6 gene signatures associated with survival of patients with melanoma and breast cancer from TCGA bulk RNA-Seq data set (SKCM and BRCA respectively). Mantel-Cox log-rank tests were used for survival analysis. *P < 0.05, ****P < 0.0001.