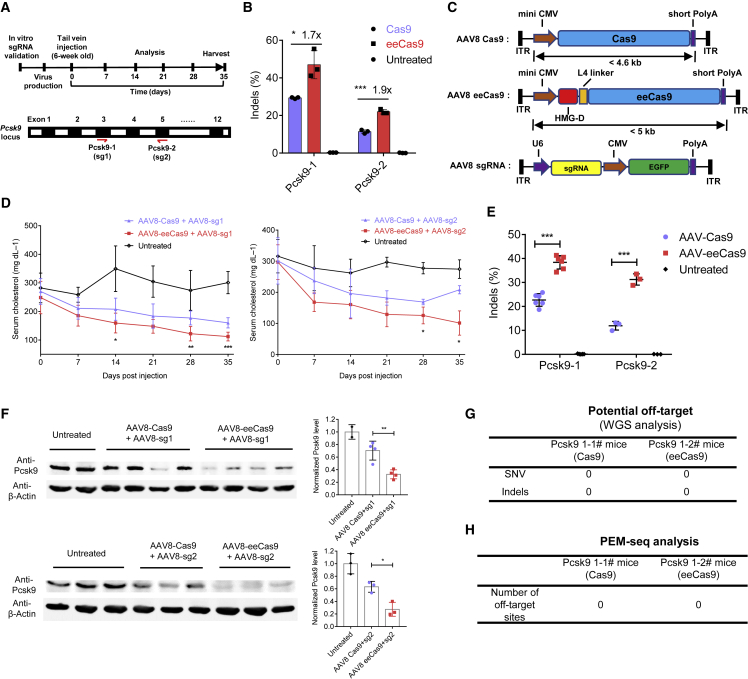
Figure 6.
AAV delivery of eeCas9 for in vivo genome editing
(A) Schematic depicting the experimental procedure to edit Pcsk9 gene in wild-type mice. Two sgRNAs (sg1 and sg2) were selected to target exon 3 and exon 5 of Pcsk9 gene, respectively. (B) sgRNA validation in NIH3T3cells via HTS analysis (n = 3 independent experiments). (C) Schematic diagram of dual AAV vector system for liver-directed gene editing. MiniCMV, minimal cytomegalovirus; short PolyA, short polyadenylation signal; ITR, inverted terminal repeat. (D) Time course of total serum cholesterol in animals treated with AAV8-Cas9 or AAV8-eeCas9. n = 6 for sg1, n = 3 for sg2. (E) Indels at Pcsk9-1(n = 6) and Pcsk9-2 (n = 3) targeting sites of Pcsk9 in mice. (F) Analysis of Pcsk9 protein expression in mouse liver by western blot. Psck9 level was determined by densitometry of western blot bands normalized by that of actin in each sample. (G) WGS analysis of potential off-target of two representative mice (1# and 2#) for Pcsk9-1 target. SNV, single-nucleotide variant; Indels, insertions and deletions. (H) PEM-seq analysis of the number of off-target sites of two representative mice (1# and 2#) for Pcsk9-1 target. Data represent means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.