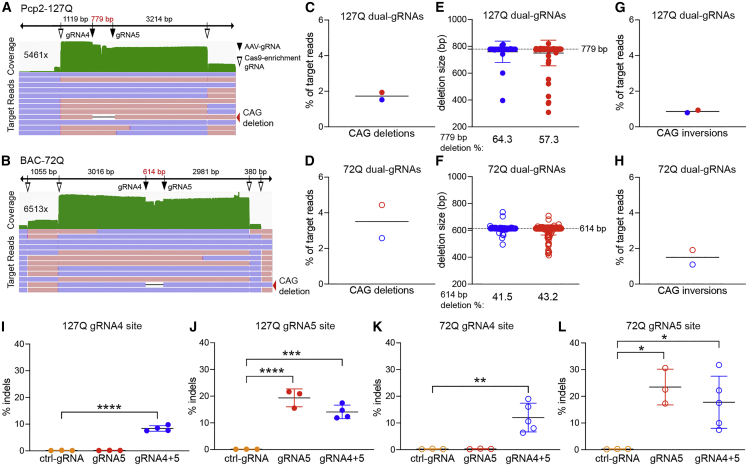
Figure 2.
Frequency of CAG deletions after dual-gRNA AAV-CRISPR editing
(A and B) Representative IGV alignment plots of nCATS reads aligned to Pcp2-127Q (A) and BAC-72Q (B) transgene ROIs. The IGV plots were cropped to highlight the relevant data, coverage is in green, maximum base coverage is on the left, positive strand reads are red, negative strand reads are blue, and reads with intended CAG deletions are shown by the red arrowheads. AAV gRNA4 and gRNA5 cleavage sites (+3-nt upstream of the PAM) are shown by black arrowheads and Cas9-enrichment gRNA sites are shown by open arrowheads; Pcp2-127Q Cas9-target enrichment with two gRNAs and BAC-72Q Cas9-target enrichment with four gRNAs. (C and D) Frequency of target reads with intended CAG deletions in Pcp2-127Q (C) (n = 2; closed circles; 4,242× and 1,843× coverage) and BAC-72Q (D) (n = 2; open circles; 4,798× and 2,520× coverage). Each point represents a single mouse and bars represent the group mean. (E and F) Distribution of intended CAG deletion sizes. Dashed line represents precise intended CAG deletion sizes of 779 bp and 614 bp in Pcp2-127Q mice (E) and BAC-72Q mice (F), respectively. Blue and red dots represent the deletion sizes in the two mice tested and the frequency of intended CAG deletions that were precise are shown below. (G and H) Frequency of target reads with inversions between the intended cleavage sites in Pcp2-127Q mice (G) (n = 2; 3,850× and 1,890× coverage) and BAC-72Q mice (H) (n = 2; 4,130× and 2,100× coverage). (I and J) Frequency of indels in Pcp2-127Q (n = 3, ctrl-gRNA and gRNA5; n = 4, gRNA4+5) at gRNA4 site (I; gRNA4+5, ∗∗∗∗p < 0.0001) and gRNA5 site (J; gRNA5, ∗∗∗∗p < 0.0001; gRNA4+5, ∗∗∗p = 0.0003) assayed by drop-off ddPCR. (K and L) Frequency of indels in BAC-72Q (n = 3, ctrl-gRNA and gRNA5; n = 5, gRNA4+5) at gRNA4 site (K) (gRNA4+5, ∗∗p = 0.0052) and gRNA5 site (L) (gRNA5, ∗p = 0.0107; gRNA4+5, ∗p = 0.0247) assayed by drop-off ddPCR. Indels were normalized to a transgene reference. Bars represent the group mean and error bars represent the standard deviation. Significance was determined by one-way ANOVA with Dunnett’s test for multiple comparisons for all ddPCR data; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.