Figure 3.
CAG repeat sizes and large deletions and rearrangements of transgenes after AAV-CRISPR editing
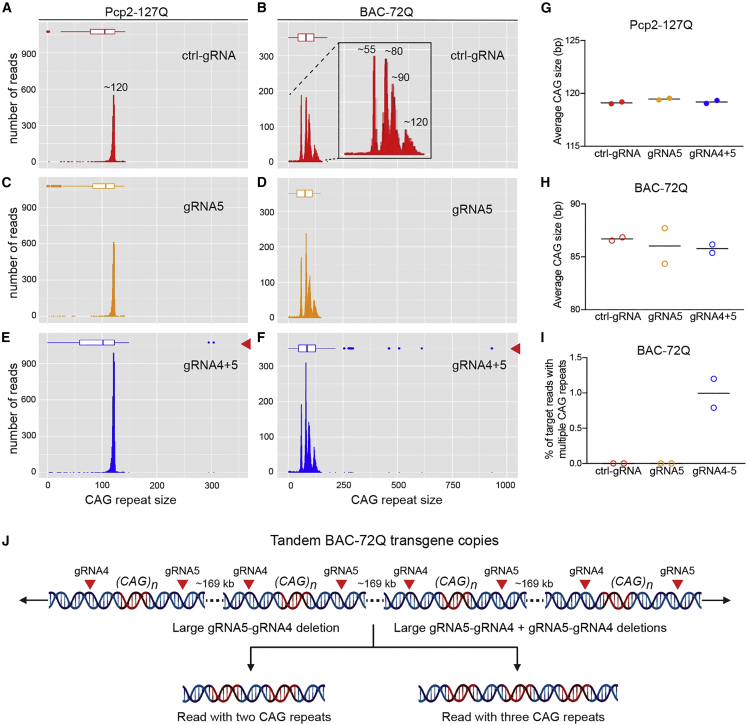
(A–F) Representative plots of CAG repeat size counts in Pcp2-127Q mice (A, C, E) and BAC-72Q mice (B, D, F); ctrl-gRNA (red) (A, B), gRNA5 (orange) (C, D), and gRNA4+5 (blue) (E, F) in nanopore Cas9-targeted reads from RepeatHMM analysis. Box plots represent CAG repeat size distribution and outliers are shown by dots. (G and H) Average CAG repeat size count for Pcp2-127Q mice (G) (n = 2, ctrl-gRNA; n = 2, gRNA5; n = 2, gRNA4+5) and BAC-72Q mice (H) (n = 2, ctrl-gRNA; n = 2, gRNA5; n = 2, gRNA4+5). Each point represents a single mouse and bars represent the group mean. (I) Frequency of target reads containing multiple CAG repeats within single reads in BAC-72Q mice (917× and 378× coverage). Bars represent the group mean. (J) Schematic of tandem transgene copies in BAC-72Q mice showing gRNA4 and gRNA5 target sites (red arrowheads) in each transgene copy. A gRNA5-gRNA4 deletion, where edit sites could be contiguous or noncontiguous, between transgene copies would yield a read containing two CAG repeats (left). Two consecutive gRNA5-gRNA4 deletions between transgene copies would cause a read containing three CAG repeats (right).