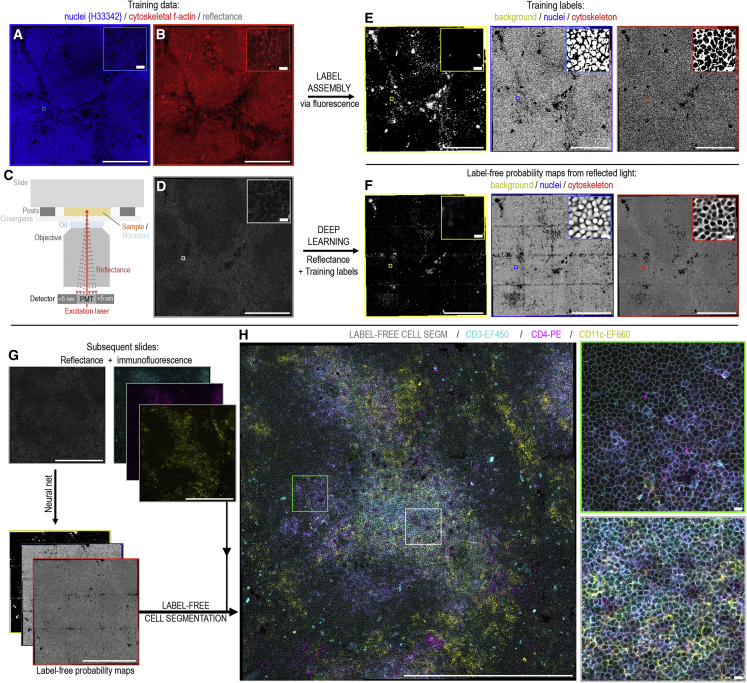
Figure 1.
Label-free cell segmentation of tissue microscopy image data collected by routine confocal microscopy
(A–D) Image data (here, mouse splenic tissue) for initial network training are obtained from serial tissue sections stained for (A) nuclei (Hoechst 33342) and (B) cytoskeletal f-actin (phalloidin-AlexaFluor 647) while simultaneously collecting (C and D) reflected laser excitation light by detector placement close (± 5 nm) to the excitation wavelength.
(E) Binary pixel-classification labels representing “background,” “nuclei,” and “cytoskeleton” classes are created by thresholding the fluorescence data.
(F) A neural network using a simple U-Net architecture is trained to output the probability that pixels in the reflectance image belong to each of these classes.
(A)–(F) show zoomed insets of the exact same image region. Comparing across these insets, the outputted probability maps (F) exhibit consistent intensities across each image field, with clear gradients that flow between the individual classifications. This enables easy, consistent instance segmentations of individual cell objects using routine watershed approaches.
(G and H) For subsequent slides, nuclei and actin stains are no longer required as the cell segmentation is achieved direct from the reflectance information via the probability map images. This establishes the cell segmentation while leaving the entire detection spectrum free for fluorescence-based analyses. For example, (H) shows the approach operating with CD3-eFluor450, CD4-PE, and CD11c-eFluor660 immunofluorescence conjugates utilizing the spectral bandwidth previously occupied by the nuclei (Hoechst 33342) and actin (phalloidin-AlexaFluor 647) stains. The label-free cell segmentation is overlaid.
(H) Insets demonstrate successful label-free cell segmentation of both CD marker-stained and entirely unstained cells in both red (green inset) and white (gray inset) pulp tissue regions.
(A–H) Main image scale bars: 250 μm, and inset image scale bars: 10 μm.