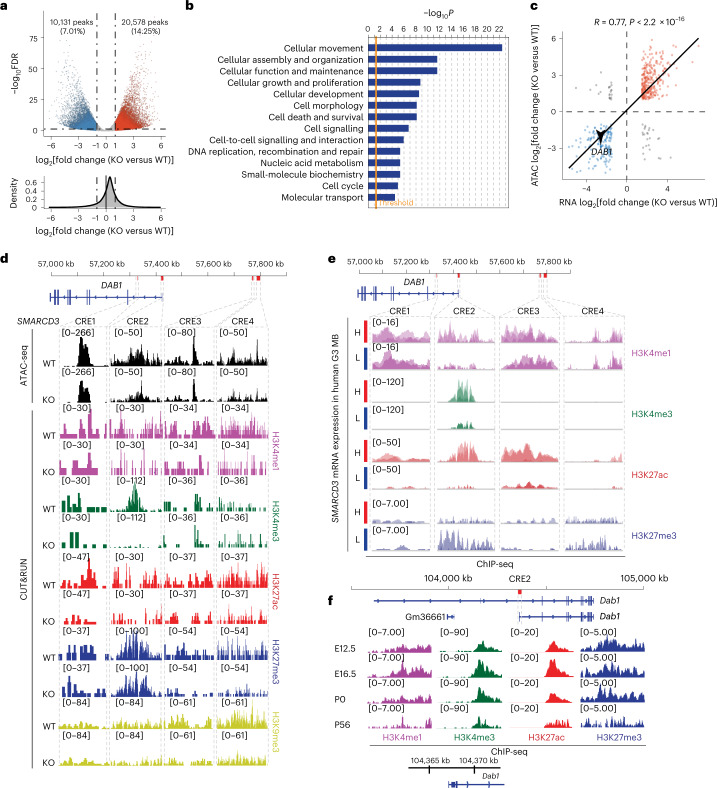
Fig. 5. SMARCD3 regulates DAB1 transcriptional activation through chromatin remodelling in MB and cerebellar development.
a, Volcano plots showing the differential accessibility (log2(fold change) in reads per peak) against the FDR (–log10) of MED8A cells with SMARCD3 KO or WT. Each dot represents one peak called by MACS3. b, The top ten molecular and cellular functions enriched according to IPA using the genes associated with reduced chromatin accessibility (FDR < 0.05; twofold change) in MED8A cells with SMARCD3 KO. c, Two-tailed Pearson’s correlation analysis of peak accessibility in ATAC-seq compared to DEGs in RNA-seq. d, ATAC-seq and histone-marker-binding signals from CUT&RUN in the DAB1 locus using MED8A cells with SMARCD3 KO or WT. The four CREs are marked by red bars and dashed-line boxes in the schematic of the genome (top). e, Histone-modification signals at the four CREs based on analyses of ChIP-seq data from five samples from patients with G3. H, high; L, low. f, Histone-modification signals at CRE2 based on analyses of ChIP-seq data from mouse cerebellum at the indicated time points. P value was calculated using right-tailed Fisher’s exact test (b).