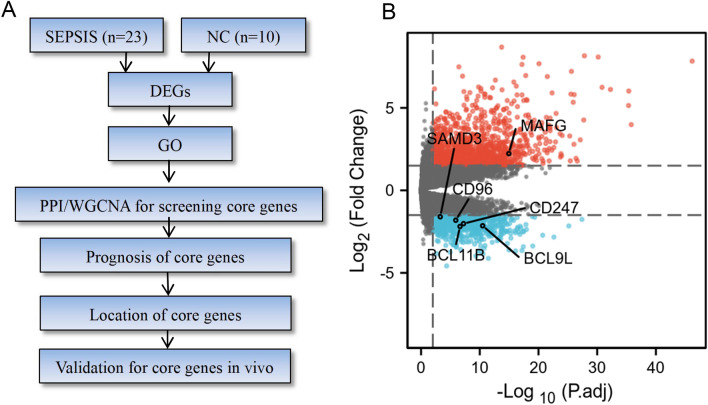
Figure 1.
Study workflow and screening of differentially expressed genes. (A): Workflow of this study. Firstly, human blood samples were collected from septic patients and healthy volunteers. RNA-seq technology and a bioinformatical approach were used for screening differentially expressed genes. Secondly, PPI analysis combined with clinical characteristics was used to make the final definition of 6 potential pivotal target genes. Lastly, single-cell sequencing was used to identify the cell lineage location of target genes. And peripheral blood sequencing from the septic mouse model was used to verify pivotal the expression profiles of key genes. (B): Volcano plot to screen differentially expressed genes. Horizontal ordinate presented as log2 (FC), each dot indicating a gene. Red color as upregulation; Blue color as downregulation. CD247, BCL9L, CD96, SAMD3 and BCL11B were downregulated in the septic group, while MAFG were up-regulated in the septic group.