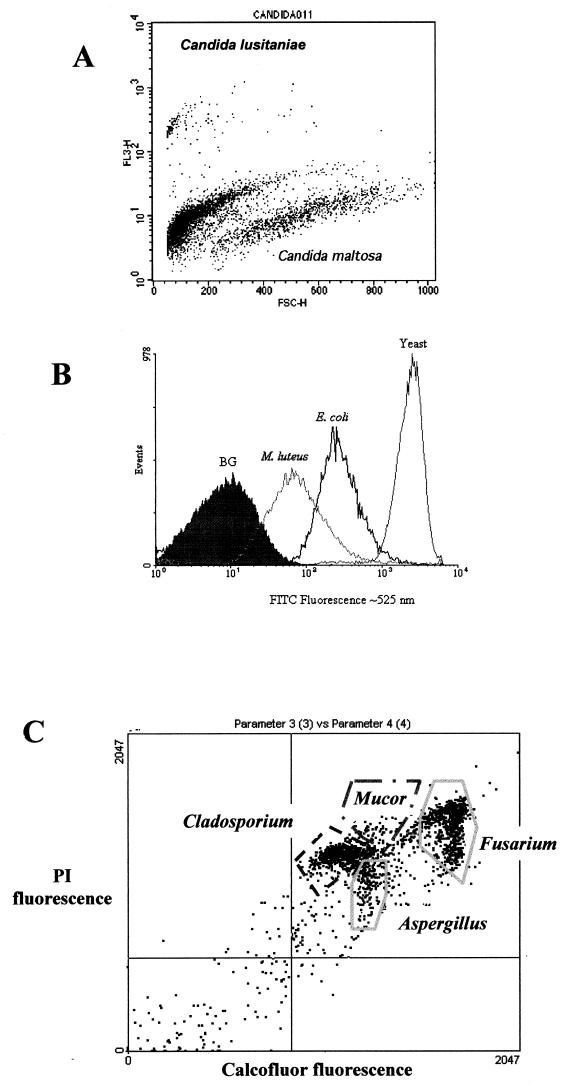
FIG. 4.
(A) Dual-parameter analysis of forward light scatter (size) and red fluorescence signals allowed the discrimination between two species of Candida, based on different fluorochrome staining backgrounds. These yeast species are indistinguishable by monoparametric analysis of forward light scatter or red autofluorescence. However, after addition of PI, they show different basal levels, and if this is plotted against size, it is possible to discriminate them. This kind of analysis permits quantification of both species in mixed cultures. (B) Quantification of different protein amounts (measured as FITC fluorescence) can be used to distinguish different microorganisms such as those represented in the histogram (from Purdue Cytometry CD-ROM, vol 2., ISSN 1091-2037, provided by Hazel M. Davey [adapted with permission of the publisher]). (C) Dual-fluorescence discrimination of fungal spores. Spores from Aspergillus, Mucor, Cladosporium, and Fusarium were fixed and stained with Calcofluor, which binds to chitin in the spore wall, and PI, which stains nucleic acids. As shown, the spores have different amounts of chitin and nucleic acids, permitting their segregation by FCM. Samples shown in panel A were run on a FACScan (Becton-Dickinson) flow cytometer, the ones shown in panel B were run on an EPICS Elite (Coulter) flow cytometer, and those shown in panel C were run on a Bryte-HS (Bio-Rad) flow cytometer.