Figure EV5. Brd4‐BD2 binds to acetylated‐EED to repress Foxp3 and Fbxw7 in Th2 cells.
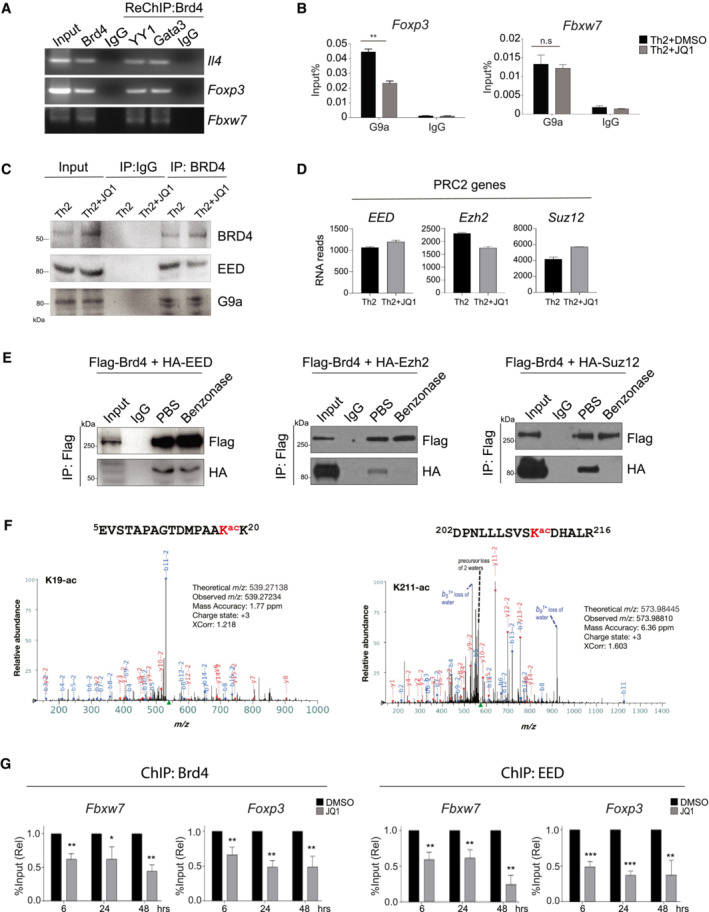
- ChIP‐PCR of the binding of Brd4 and Re‐ChIP‐PCR of YY1 and Gata3 on the gene loci of Il4, Foxp3, and Fbxw7 in mouse Th2 cells.
- ChIP‐qPCR analysis of the binding of G9a on the gene loci of Foxp3 and Fbxw7 in mouse Th2 cells treat with or without JQ1 (250 nM).
- Th2 cells were differentiated for 6 days, treated with JQ1 (250 nM) on Day 0. Lysates are immunoprecipitated with BRD4 followed by Western blotting analysis of BRD4, EED, and G9a.
- RNA‐seq data analysis showing expression of PRC2 key genes in mouse Th2 cells treated with or without JQ1 (250 nM). Data are representative of two biological replicates.
- HEK293T cells are transfected with Flag‐Brd4 and HA‐EED (left), HA‐Ezh2 (middle), or HA‐Suz12 (right) plasmids. Lysates are immunoprecipitated with Flag‐tagged Brd4, treated in vitro with PBS and Benzonase, followed by Western blotting of HA to detect HA‐EED, HA‐Ezh2, and HA‐Suz12.
- HEK293T cells are transfected with Flag‐Brd4 and HA‐EED plasmids. Lysates are immunoprecipitated with HA, followed by liquid chromatography with tandem mass spectrometry (LC–MS/MS) analysis. MS/MS spectrum for the identified acetylated peptides containing Lysine acetylation at position K19 and K211. Peak heights show the relative abundance of the corresponding fragmentation ions. The peptide sequences are shown at the top of the corresponding MS/MS spectrum with acetylated lysine residues highlighted in red. The identified fragmentation y (red color) and b (blue color) ions are indicated.
- Th2 cells were differentiated for 3 days and then treated with DMSO and JQ1 (250 nM) for 6, 24 and 48 h, followed by ChIP‐qPCR analysis of binding of Brd4 and EED on Foxp3 and Fbwx7.
Data information: Mouse naïve CD4+ T cells were cultured in Th2 polarization condition and treated with or without inhibitors on Day 0 and were differentiated for 6 days before analysis, unless otherwise specified. All Western blotting data are representative of three independent experiments. All ChIP‐qPCR data represent mean ± SD and are representative of three independent experiments. Data are analyzed by Paired t test. *P < 0.05; **P < 0.01; and ***P < 0.001.
Source data are available online for this figure.
