Figure 5. Adipocyte autophagy loss activates NRF2‐EPHX1 pathway and alters intratissual oxylipin balance.
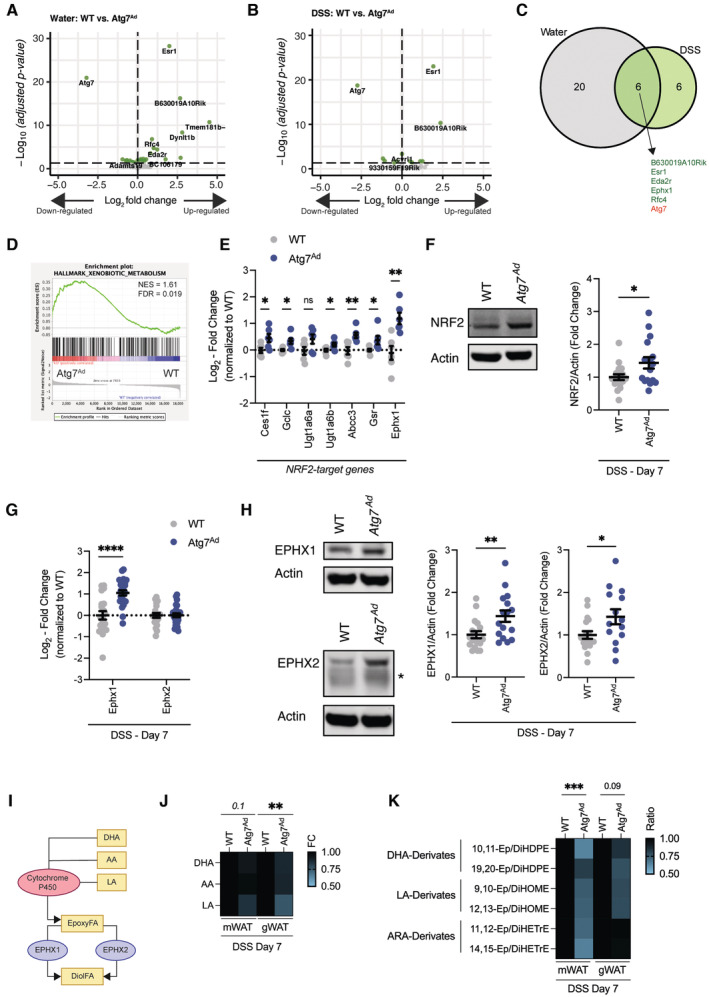
- Differential gene expression in visceral adipocytes from water‐treated WT and Atg7 Ad animals 2 weeks after tamoxifen treatment.
- Differential gene expression in visceral adipocytes from DSS‐treated WT and Atg7 Ad animals on day 7 post‐DSS treatment.
- Venn diagram of commonly regulated genes between Atg7‐deficient and Atg7‐sufficient adipocytes during water or DSS treatment.
- GSEA enrichment analysis between Atg7‐deficient and Atg7‐sufficient adipocytes during DSS treatment.
- Fold change expression of NRF2‐target genes in primary visceral adipocytes on day 7 after DSS induction from normalized counts of RNAseq dataset (n = 6/group).
- Representative immunoblot for NRF2 protein expression and quantification (n = 16–18/group).
- Transcriptional expression of Ephx1 and Ephx2 in visceral adipocytes on day 7 after DSS induction (n = 20–25/group).
- Representative immunoblot of EPHX1 and EPHX2 in gonadal adipose tissues on day 7 after DSS induction. Asterix indicating nonspecific band (n = 14–18/group).
- Schematic overview of cytochrome P450‐EPHX oxylipin pathway.
- Normalized fold change differences in epoxy fatty acid precursor fatty acids, docosahexaenoic acid (DHA), arachidonic acid (AA), and linoleic acid (LA) in mWAT and gWAT (n = 13–14/group).
- Normalized ratios of epoxy fatty acid to their corresponding diol fatty acid pairs in mWAT and gWAT (n = 6–8/group).
Data are represented as mean ± s.e.m. (E–H, L) Unpaired Student's t‐test. (J, K) Two‐way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Source data are available online for this figure.
