Fig. 1 |. CTCF depletion leads to widespread upregulation of antisense transcription at divergent promoters.
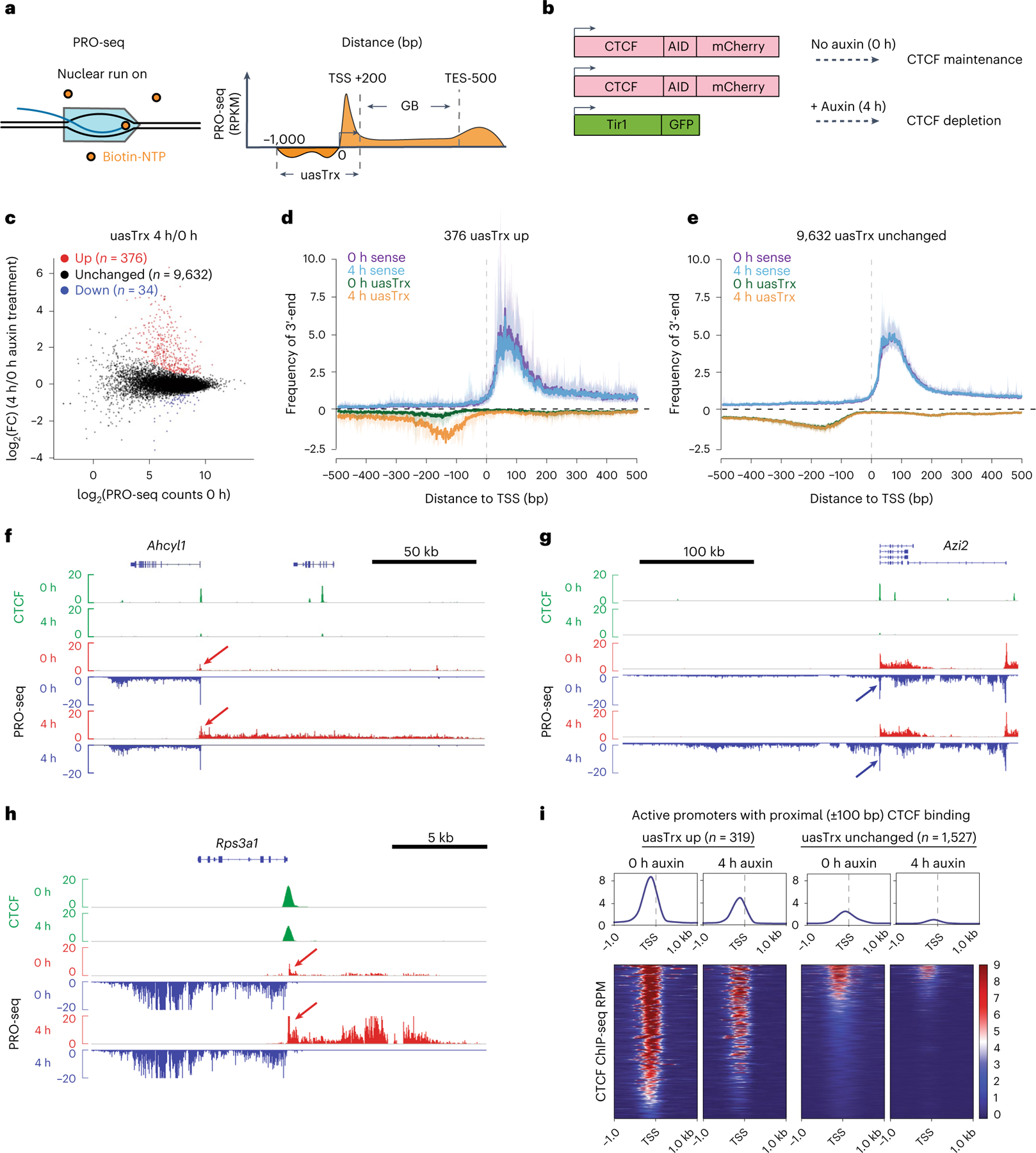
a, Schematic of the PRO-seq experiment (left) and quantification strategy (right). b, Schematic of the experimental set-up. c, PRO-seq MA plot of control versus CTCF-depleted cells on the antisense strand (−1,000 base pair (bp) to +200 bp relative to annotated TSS) in G1E-ER4s. Differentially expressed transcripts are highlighted in color. d, Metaplot of sense and antisense 3′-end PRO-seq mapping, centered at annotated TSSs and plotted with respect to sense orientation for genes with upregulated uasTrx. Solid lines and shading show average signals and the 12.5/87.5th percentiles, respectively. e, As in d for unchanged uasTrx genes. f, Genome browser views of CTCF ChIP-seq (green) and PRO-seq signals (plus strand in red, minus strand in blue) at the Ahcyl1 locus. Arrows point to increased uasTrx. g, As in f for the Azi2 locus. h, As in f for the Rps3a1 locus. i, Heatmaps showing CTCF occupancy at active promoters with proximal (±100 bp) CTCF-binding (up, n = 319; unchanged, n = 1,527) sorted by occupancy level, and shown with respect to sense orientation.
