Fig. 2 |. CTCF inhibits uasTrx directly and proximally, and independently of its architectural functions.
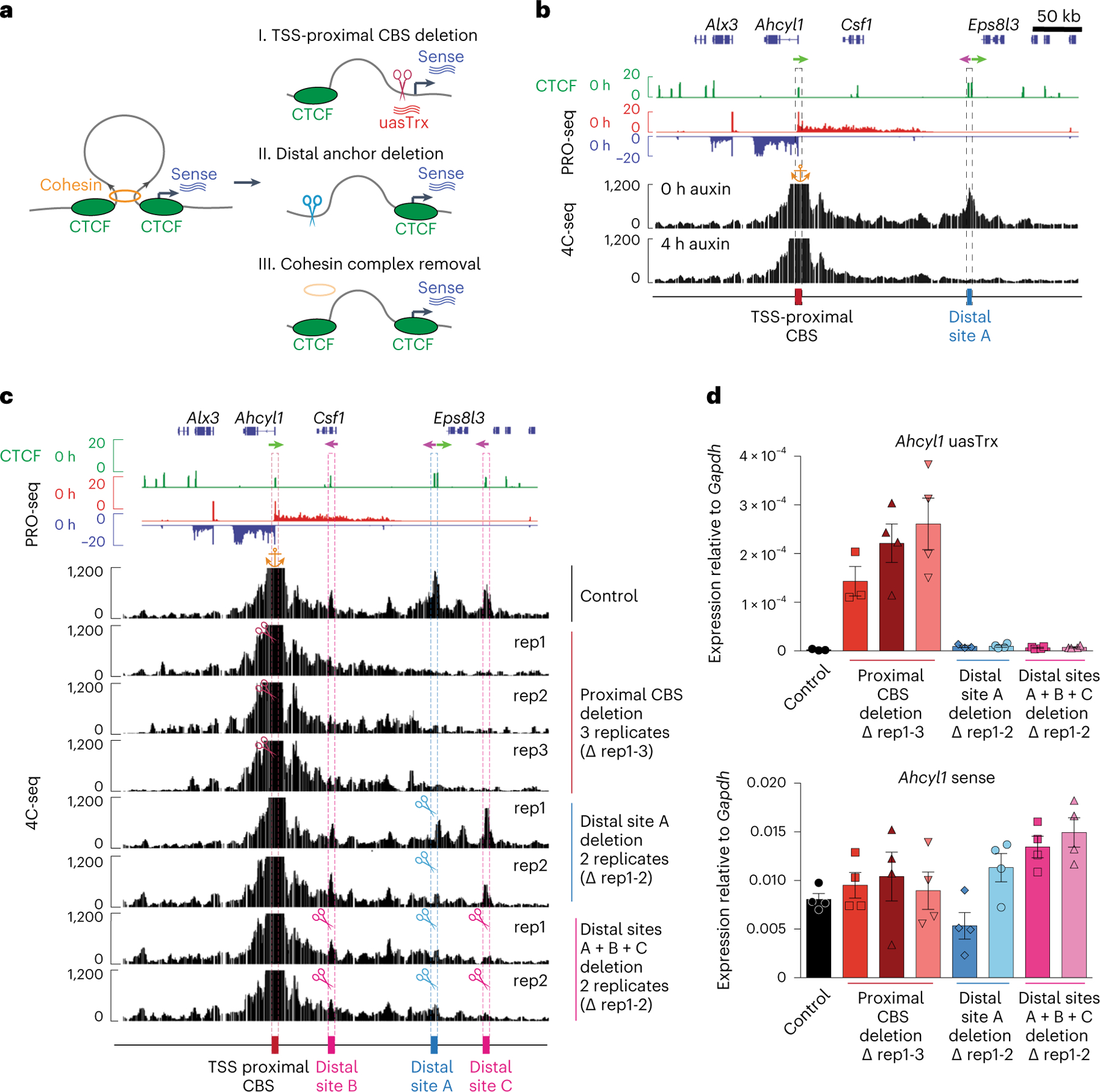
a, Illustration of the experimental strategy and summarized findings from this figure and Extended Data Figs. 3–5. b, Genome browser views of CTCF ChIP-seq, PRO-seq and 4C-seq signals at Ahcyl1. Arrows indicate CTCF motif orientation. 4C-seq anchored at the Ahcyl1 promoter with (4 h auxin) and without (0 h auxin) CTCF degradation. The orange anchor indicates the 4C-seq viewpoint. Sites of interest are indicated below the track and are highlighted by dashed boxes. c, Genome browser tracks of CTCF ChIP-seq and PRO-seq and representative 4C-seq profiles of Ahcyl1 control and edited clones. Similar observations were made in two or three independent 4C-seq experiments. The orange anchor indicates the 4C-seq viewpoint. Arrows indicate CTCF motif orientation. Scissors indicate CRISPR/Cas9-edited regions. d, RT–qPCR of Ahcyl1 uasTrx and sense transcription in control and edited clones. Transcripts were normalized to Gapdh (error bar indicates s.e.m.; n = 4, except for uasTrx control, proximal and distal CBS deletion rep1, for which n = 3). Same analyses with different primer pairs are depicted in Extended Data Fig. 4b,c.
