Extended Data Fig. 1 |. CTCF depletion leads to widespread uasTrx upregulation at divergent promoters.
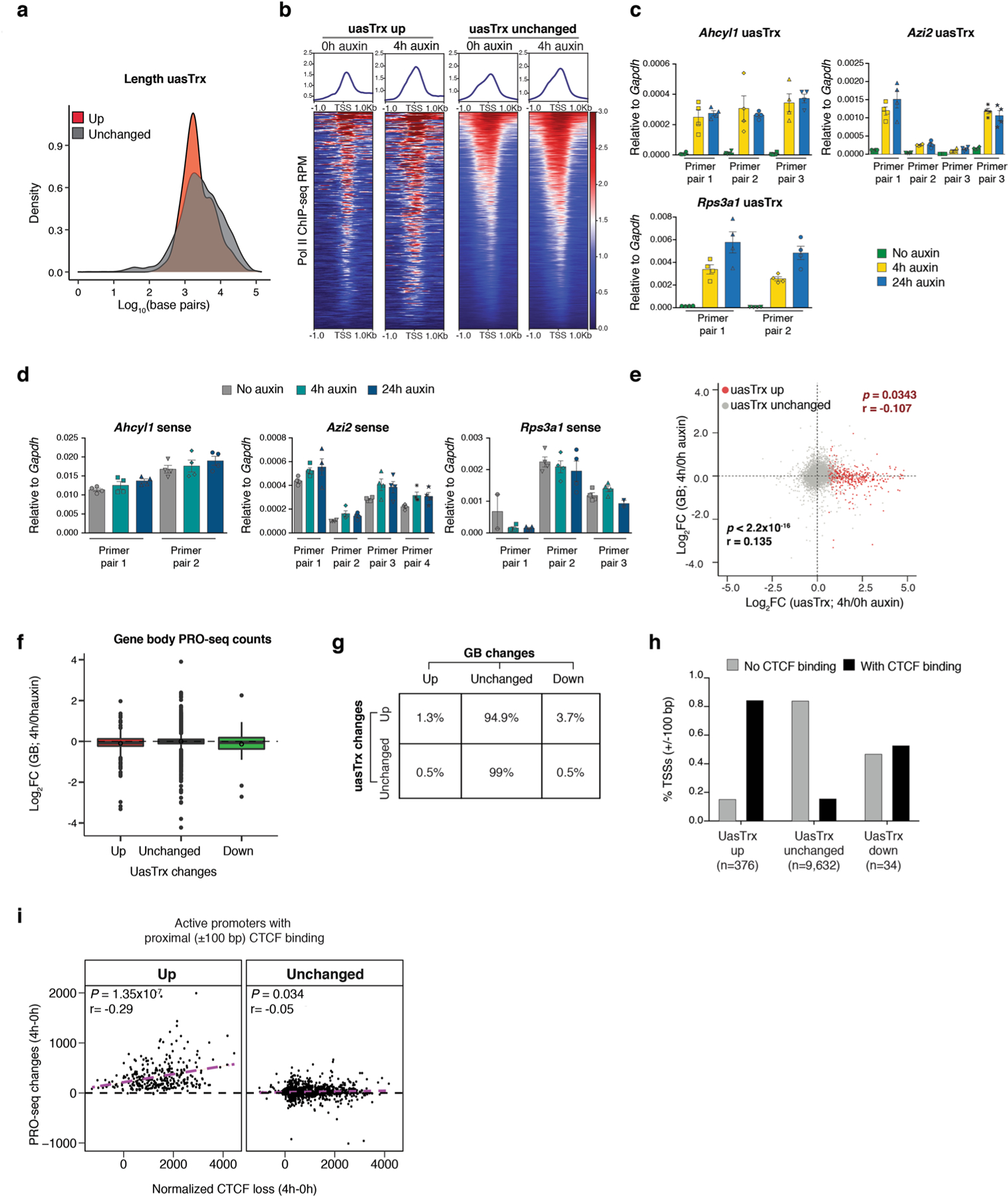
a, Distribution of uasTrx lengths, grouped by changes in response to CTCF depletion. b, Row-linked heatmaps showing Pol II occupancy at active promoters, grouped by antisense changes (up, n = 376; unchanged, n = 9,632) upon CTCF depletion, sorted by occupancy level, and shown with respect to sense orientation. c, RT–qPCR of uasTrx for Ahcyl1 at indicated time points after CTCF depletion. Transcripts were normalized to Gapdh (error bar: SEM; n = 4). d, same as (c) but quantifying nascent sense transcripts. e, Scatterplot comparing transcriptional changes in gene body (GB) versus uasTrx. Data points grouped and colored based on uasTrx changes. P values were calculated by Spearman rank correlation test, r is the correlation coefficient. f, Log-transformed PRO-seq fold changes in GB after CTCF depletion, grouped by uasTrx changes. Lower and upper box ends represent the first and third quartiles with the median indicated as a horizontal line within the box. Mean is indicated by a circle within the box. Whiskers define the smallest and largest values within 1.5 times the interquartile range below the first or above the third quartile, respectively. Outliers are plotted as individual dots. g, Transcriptional changes in uasTrx and GB after CTCF depletion. h, Percentage of promoters with and without proximal (±100 bp) CBSs as a function of uasTrx changes. i, Correlation between PRO-seq changes and CTCF loss at uasTrx with proximal (±100 bp) CTCF binding. Linear regression line shown in magenta. P values were calculated by Spearman rank correlation test, r is the correlation coefficient.
