Extended Data Fig. 2 |. CTCF depletion in human HCT-116 and mESCs leads to antisense transcriptional changes.
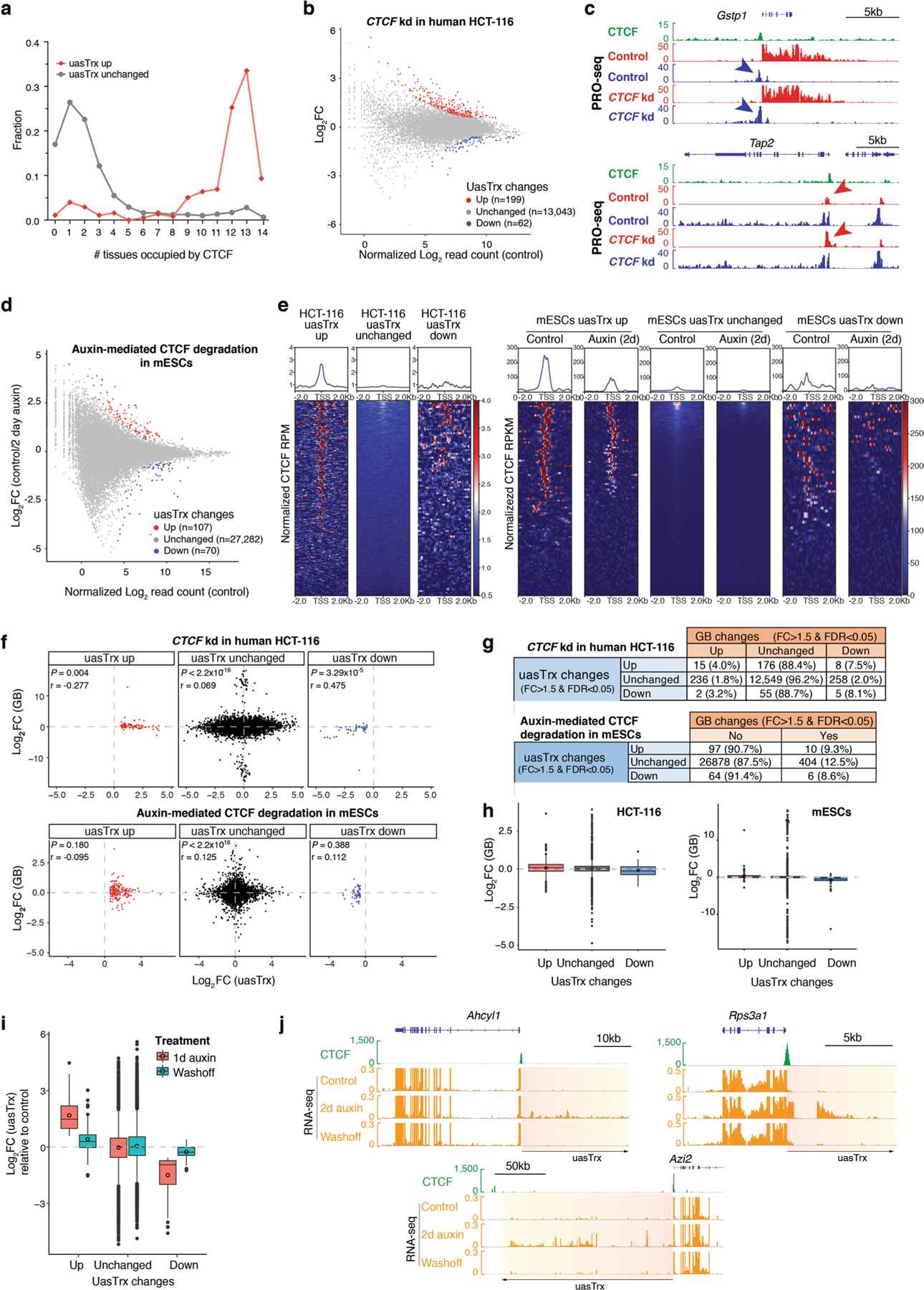
a, Fraction of TSSs detected in the indicated numbers of mouse tissues where CTCF binds in proximity (within ± 100 bp), grouped by uasTrx changes. b, PRO-seq MA plot of control versus CTCF-depleted cells on the antisense strand (−1000 bp to +200 relative to annotated TSS) in human HCT-116 cells. Differentially expressed transcripts highlighted in color. c, Browser views of CTCF ChIP-seq (mm9 liftover from Rao et al., 2014) and PRO-seq signals at Gstp1 and Tap2 loci in HCT-116 cells. Arrows highlight location of CTCF-repressed uasTrx. Arrow color indicates uasTrx strandedness. kd, knockdown. d, RNA-seq MA plot of control versus CTCF-depleted cells on the antisense strand (−1000 bp to +200 relative to annotated TSS) in mESCs. Differentially expressed transcripts highlighted in color. e, Row-linked heatmaps showing CTCF occupancy at active promoters, grouped by uasTrx changes, sorted by binding enrichment levels, and shown with respect to sense orientation in HCT-116 cells and mESCs. f, Correlation between uasTrx and GB changes after CTCF depletion in PRO-seq data from HCT-116 cells, and RNA-seq data from mESCs. P value was calculated by Spearman rank correlation test; r is the correlation coefficient. g, Transcriptional changes in uasTrx and GB after CTCF depletion in PRO-seq from HCT-116 cells and RNA-seq data from mESCs. h, Log-transformed PRO-seq and RNA-seq fold changes in GB after CTCF depletion in HCT-116 cells and mESCs, respectively. Lower and upper box ends represent the first and third quartiles with the median indicated as a horizontal line within the box. Mean is indicated by a circle within the box. Whiskers define the smallest and largest values within 1.5 times the interquartile range below the first or above the third quartile, respectively. Outliers are plotted as individual dots. i, Log-transformed RNA-seq fold change in uasTrx in indicated conditions over control in mESCs. j, Brower views of CTCF ChIP-seq and RNA-seq signals at Ahcyl1, Azi2 and Rps3a1 loci in mESCs. Orange to yellow boxes and black arrow indicate (direction of) uasTrx.
