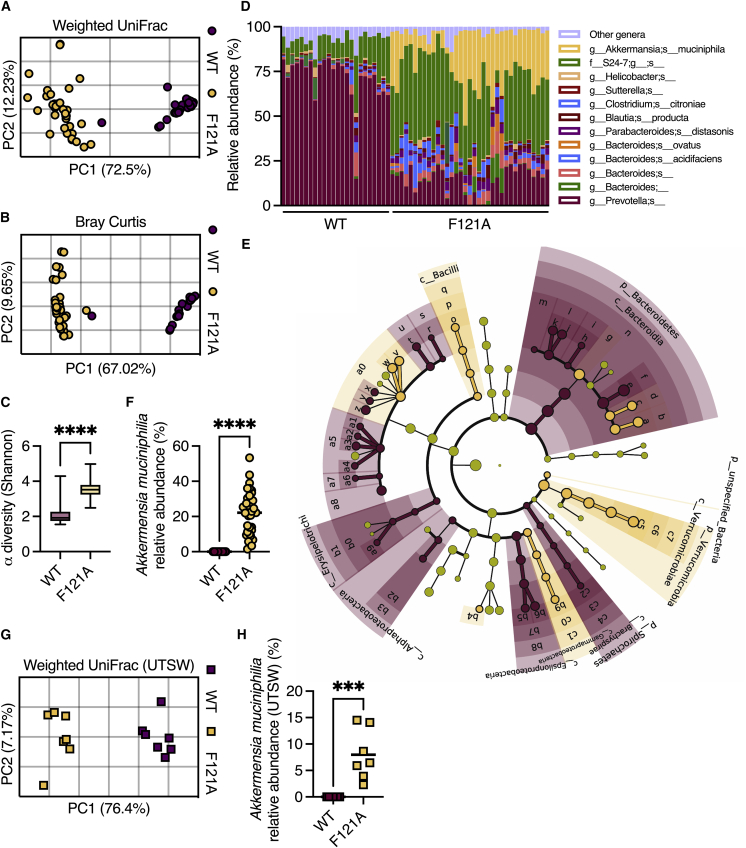
Figure 5.
Becn1F121A mice contain an altered gut microbiota
16S rRNA sequencing was performed to characterize gut microbiota composition.
(A and B) (A) PCoA of fecal microbiota β diversity based on weighted UniFrac and (B) Bray-Curtis dissimilarity in mice housed at the Bar-Ilan University.
(C) α diversity comparison based on richness and evenness.
(D) Relative taxonomic composition.
(E) Cladogram depicting LEfSe analysis of differently abundant bacteria in wild-type and Becn1F121A mice.
(F) Relative abundance of Akkermansia muciniphila in mice housed at the Bar-Ilan University.
(G) Principal coordinates analysis (PCoA) of fecal microbiota β diversity based on weighted UniFrac on mice housed at UTSW.
(H) Relative abundance of Akkermansia muciniphila in mice housed at UTSW.
(A, B, and F–H) Each symbol represents a mouse. (D) Each column represents a mouse. ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; (C, F and H) Student’s t test. WT, wild type; F121A, Becn1F121A; UTSW, UT Southwestern.