Abstract
Immune checkpoint inhibitors (ICIs) enhance anticancer immunity by releasing repressive signals into tumor microenvironments (TMEs). To be effective, ICIs require preexisting immunologically “hot” niches for tumor antigen presentation and lymphocyte recruitment. How the mutational landscape of cancer cells shapes these immunological niches remains poorly defined. We found in human and murine colorectal cancer (CRC) models that the superior antitumor immune response of mismatch repair (MMR)–deficient CRC required tumor cell–intrinsic activation of cGAS-STING signaling triggered by genomic instability. Subsequently, we synthetically enforced STING signaling in CRC cells with intact MMR signaling using constitutively active STING variants. Even in MMR-proficient CRC, genetically encoded gain-of-function STING was sufficient to induce cancer cell–intrinsic interferon signaling, local activation of antigen-presenting cells, recruitment of effector lymphocytes, and sensitization of previously “cold” TMEs to ICI therapy in vivo. Thus, our results introduce a rational strategy for modulating cancer cell–intrinsic programs via engineered STING enforcement to sensitize resistant tumors to ICI responsiveness.
Enforced STING signaling in tumor cells reprograms the TME and sensitizes “cold” tumors to checkpoint inhibitor therapy.
INTRODUCTION
Immune checkpoint inhibitors (ICIs) have revolutionized cancer therapy and brought benefits to many tumor patients that were inconceivable 15 years ago (1). ICIs relieve inhibitory signals (checkpoints) from receptors such as PD1 or CTLA4 on tumor-infiltrating lymphocytes (TILs), which are frequently usurped by cancer cells, and thereby restore antitumor immunity via CD8+ cytotoxic T lymphocytes (CTLs) (2). However, a substantial fraction of cancer patients are resistant to ICI therapy because their T cells fail to infiltrate the tumor microenvironment (TME) or lack an initial activating signal in response to tumor-associated antigens in the vicinity of cancer cells, which are provided by antigen-presenting cells (APCs). The sensitivity of malignant tumors to ICI therapy is therefore critically dependent on the initial recruitment, activation, and/or differentiation of lymphoid and myeloid immune cells into intratumoral inflammatory and immunologically “hot” niches. Because the biology of these niches is typically determined by the mutational landscape and the aberrant molecular signaling within the cancer cell itself (3), understanding and engagement of these mechanisms may offer novel perspectives to develop TME-modulating strategies that can overcome the current limitations of ICI treatment.
To date, three predictive biomarkers for anti-PD1 or anti-PDL1 ICI responses have been validated: the mismatch repair (MMR) status of the cancer cell (4), the tumor mutational burden (TMB) (5), and the level of PDL1 expression in the tumor tissue (6). Deficiency in DNA damage repair genes due to the loss of function of elements of the MMR machinery, such as MutL homolog 1 (MLH1), MutS homolog 2 (MSH2), DNA polymerase delta 1 (POLD1), and DNA polymerase epsilon (POLE) (7, 8), results in genomic instability, a cancer hallmark (9), and a hypermutator phenotype with accumulation of unrepaired replication-associated errors, numerous alterations in microsatellite sequence, a phenomenon called microsatellite instability (8, 10), and a genome-wide increase in somatic mutations with an elevated TMB and enrichment of neoantigens in the tumor tissue (11, 12).
In colorectal cancer (CRC), which is one of the most prevalent malignancies in the world, with nearly 1 million deaths in 2020 (13, 14), approximately 15 to 20% of tumors harbor inherited or sporadic loss-of-function mutations in or epigenetic silencing of MMR regulators (12). These MMR-deficient (dMMR) tumors generally have a better clinical prognosis than microsatellite-stable CRCs (15, 16), and they are particularly responsive to ICI immunotherapy (4, 17, 18). However, approximately 50% of dMMR tumors do not respond to immune checkpoint blockade, suggesting that the neoantigen-generated mutation burden is not sufficient to drive responsiveness to immunotherapy.
Using murine and human CRC models, we observed that deficiency in MMR genes triggered an aberrant accumulation of DNA in the cytosol that subsequently activated the cGAS-STING innate immune pathway within the tumor cell, in addition to genomic mutations. These events shaped an immunologically active TME with TIL recruitment and productive antitumor immunity in vivo. Building on these observations, we engineered constitutively active gain-of-function STING variants to enforce tumor cell–intrinsic STING pathway activation. Even in MMR-proficient cancer cells, this strategy was sufficient to induce local APC activation in vivo, recruitment of lymphoid effector cells, and sensitization of previously “cold” tumors to ICI therapy. Together, these results present a synthetic strategy to enforce STING signaling within cancer cells to reprogram TMEs for responsiveness to ICI therapy.
RESULTS
MMR deficiency drives IFN signaling in CRC
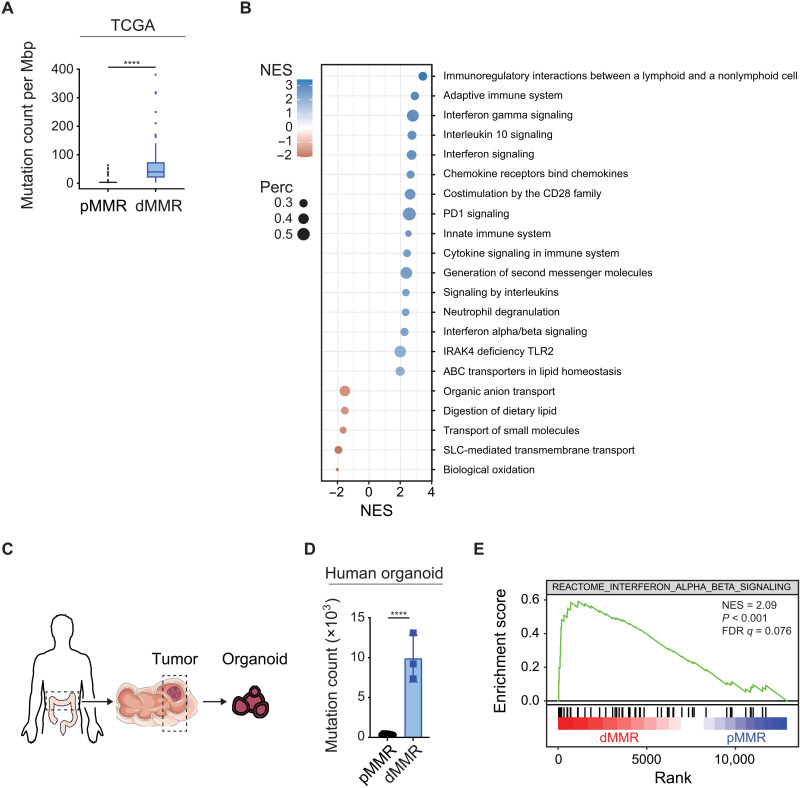
To identify immunostimulatory pathways in dMMR CRCs that could be responsible for their superior susceptibility to ICIs, we first retrieved genome and RNA-seq data from 524 colorectal and rectal adenocarcinoma patient samples from The Cancer Genome Atlas (TCGA). On the basis of the mutational status of MLH1, MSH2, POLD1, and POLE, which are frequently inactivated in CRC (7, 8), we divided the samples into dMMR tumors (n = 73) and MMR-proficient (pMMR) tumors (n = 451). Consistent with previous data (10), the frequency of genomic mutations per megabase (TMB) was significantly higher in dMMR than in pMMR cases (Fig. 1A). Next, we performed a preranked gene set enrichment analysis (GSEA) of differentially expressed genes between these groups using the Reactome pathway database. The top differentially regulated pathways between dMMR and pMMR tumors involved immune and inflammatory processes with a prominent representation and enrichment of interferon (IFN) signaling–related signatures such as “IFN-γ signaling,” “IFN signaling,” and “IFN-α/β signaling” in the dMMR cancers (Fig. 1B). Thus, dMMR in CRC is associated not only with an increased TMB but also with activation of IFN signaling in cancer tissue.
Fig. 1. MMR deficiency drives IFN signaling in CRC.
Exome and RNA-seq data of pMMR and dMMR tumors (A and B) from 524 colorectal and rectal adenocarcinoma patients (TCGA) or (D and E) from patient-derived primary organoids. (A) Mutation count per Megabase pairs (Mbp) of pMMR versus dMMR tumors. (B) GSEA of differentially expressed gene sets from the Reactome database comparing pMMR versus dMMR tumors (+ve NES: dMMR). (C) Schematic representation of organoids from patients. (D) Mutation count of pMMR (n = 9) and dMMR (n = 3) tumor organoids. (E) GSEA of RNA-seq data using the Reactome IFN-α/β signaling gene set comparing dMMR (red) versus pMMR (blue). Student’s t test (A and D) was used to determine significance.
The TCGA datasets were retrieved from tumor tissue blocks that contained epithelial cancer cells, stroma, and infiltrating immune cells. To selectively explore putative cancer cell–intrinsic inflammatory signals in primary human CRC specimens, we next prepared tumor organoids from CRC patients for whole-exome sequencing (WES), RNA sequencing (RNA-seq), and GSEA (Fig. 1C). In our biobank, we identified three dMMR organoids with mutations in MSH2 or POLE that were compared to nine pMMR samples without mutations in known MMR genes (tables S1 and S2). In these dMMR tumor organoids, we observed increased TMB (Fig. 1D). In addition, type I IFN signaling represented by IFN-α/β signaling was one of the most prominently enriched pathways in the dMMR cases (Fig. 1E and fig. S1A), demonstrating tumor cell–intrinsic activation of these immune signaling pathways.
MMR deficiency triggers tumor cell–intrinsic STING pathway activation
To dissect the mechanisms that drive tumor cell–intrinsic IFN signaling in dMMR CRC, we used a genetically tractable murine model and edited the MC38 CRC cell line using CRISPR-Cas9. First, we homozygously disrupted MLH1 in MC38 cells (Fig. 2A). Subsequent RNA-seq and GSEA confirmed a strong induction of IFN signaling upon MLH1 deletion, which was represented by the top differentially regulated signatures IFN signaling and IFN-α/β signaling (Fig. 2B). In addition, in the cytosol of MLH1−/− tumor cells, we detected an increase in genomic DNA (fig. S2C), which is released from the nucleus upon continuous DNA damage (19–21).
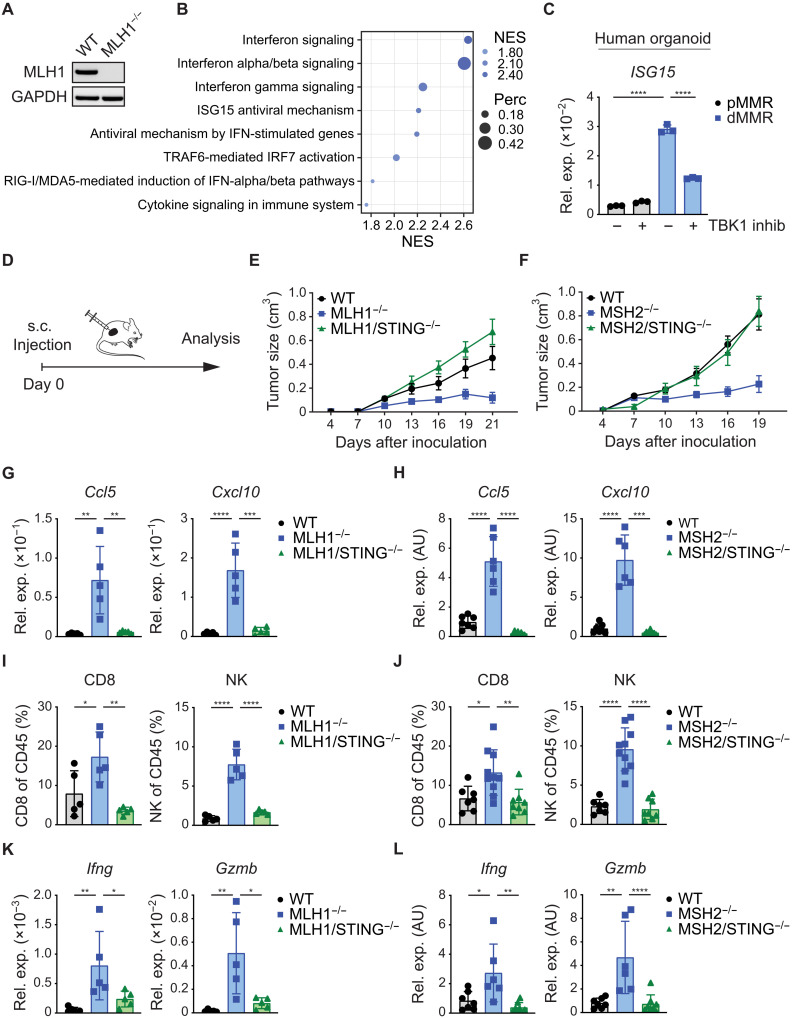
Fig. 2. STING signaling in dMMR CRC mediates immunogenicity.
(A) MLH1 deletion in MC38 tumor cells was confirmed by Western blotting. (B) GSEA of RNA-seq data to identify differentially expressed gene sets of WT versus MLH1−/− MC38 tumor cells by using the Reactome database (+ve NES: MLH1−/−). (C) The relative gene expression of ISG15 in pMMR (PDO#12) versus dMMR (PDO#7) cultured primary organoids was quantified by qPCR 16 hours after TBK1 inhibitor treatment. (D) Schematic representation of the experimental setup in vivo. s.c. subcutaneous. (E) Growth of subcutaneously inoculated WT, MLH1−/−, or MLH1/STING−/− MC38 tumor cells (n = 5) or (F) WT, MSH2−/−, or MSH2/STING−/− MC38 tumor cells (n = 7 to 10) in syngeneic WT C57Bl/6 mice. (G to L) At the end point (MLH1 day 21; MSH2 day 19), subcutaneously grown tumors were explanted for FACS and qPCR analysis. (G and H) Relative gene expression of chemokines (Ccl5, Cxcl10) was quantified by qPCR. (I and J) FACS analyses displaying the percentages (CD8, NK) of live/CD45+ cells. (K and L) The relative gene expression of cytotoxic effector molecules (Gzmb, Ifng) was quantified by qPCR. The data represent n = 3 technical replicates (C). The data are presented as the mean ± SEM (E and F). One-way ANOVA (C and G to L) was used to determine significance. AU, arbitrary units.
Mammalian cells detect cytosolic DNA via the DNA sensor cGAS (22), which subsequently generates the second messenger cGAMP to activate the innate immune adapter molecule STING, leading to TBK1 signaling and transcription of type I IFN genes (23). To explore the role of the cGAS-STING pathway in dMMR-triggered IFN signaling, we created double-mutated MC38 cells with codeletion of MLH1 and cGAS (MLH1/cGAS−/−) or MLH1 and STING (MLH1/STING−/−) (fig. S2A). Deletion of MLH1 led to cell-autonomous production of cGAMP in a cGAS-dependent manner (fig. S2D). Moreover, MLH1 deficiency resulted in increased IFN-β production, which was also strictly dependent on cGAS-STING signaling (fig. S2E). In line with the increased IFN-β production, MLH1-deficient MC38 cells also displayed increased signal transducer and activator of transcription 1 (STAT1) phosphorylation, which mediates signals downstream of the type I IFN receptor IFN-α and β receptor subunit 1 (IFNAR1) (fig. S2I).
Next, we measured the expression of the IFN-stimulated gene (ISG) Isg15 as a prototypical marker for productive IFN signaling (24) and the expression of Ccl5 and Cxcl10, which are critical TIL-attracting chemokines (25). Isg15, Ccl5, and Cxcl10 were strongly induced upon MLH1 deletion, and this induction was dependent on cGAS and STING (fig. S2J). To create an additional model of dMMR, we deleted MSH2 in MC38 cells without or with cGAS or STING codeletion (fig. S2B). Similar to MLH1 disruption, MSH2 deficiency increased the presence of cytosolic DNA (fig. S2F), cGAS-dependent cGAMP production (fig. S2G), cGAS- and STING-mediated IFN-β production (fig. S2H), STAT1 signaling (fig. S2K), and ISG expression (fig. S2K).
To test the role of autocrine type I IFN signaling, we next blocked the type I IFN receptor IFNAR1 in MLH1- or MSH2-deficient MC38 tumor cells with anti-IFNAR1 antibodies. Blocking IFNAR1 reduced the expression of Isg15, Ccl5, and Cxcl10, which indicates that IFNs produced by dMMR MC38 tumor cells drive autocrine transcription of ISGs at least in part via IFNAR1 (fig. S3, A and B). Together, these experimental data demonstrate that defects in the MMR machinery lead to an accumulation of DNA in the cytosol, which stimulates the production of cGAMP via cGAS to engage STING for the activation of type I IFN gene transcription.
To translate these findings to human CRC, we used our patient-derived organoid models. In the dMMR organoids, we observed enhanced ISG15 gene expression in comparison to the pMMR counterparts. This ISG expression was significantly reduced upon pharmacological TBK1 inhibition (Fig. 2C), indicating that the principal mechanisms of colon cancer cell–intrinsic dMMR-triggered ISG induction are conserved between mice and humans.
STING signaling in dMMR CRC mediates immunogenicity
Next, we studied the consequences of tumor cell–intrinsic STING activation on tumor immune TMEs and dMMR cancer cell growth in vivo. To this end, we subcutaneously transplanted wild-type (WT), MLH1- and MLH1/STING-deficient, or MSH2- and MSH2/STING-deficient MC38 tumor cells into syngeneic C57BL/6 mice (Fig. 2D). While WT, MLH1- and MLH1/STING-deficient, or MSH2- and MSH2/STING-deficient MC38 tumor cells proliferated equally in in vitro cultures (fig. S3, C and D), MLH1- and MSH2-deficient MC38 cells grew smaller tumors in vivo (Fig. 2, E and F, and fig. S3, E and F). This reduced tumor growth was associated with up-regulated expression of the chemokines Ccl5 and Cxcl10, which are critical for the recruitment of cytotoxic CD8+ T cells and natural killer (NK) cells into tumor tissues (Fig. 2, G and H) (25). Consistently, we also detected higher frequencies of both CTLs and NK cells in the TMEs of MLH1- or MSH2-deficient tumor cells (Fig. 2, I and J) and up-regulation of the cytotoxic effector molecules granzyme B (Gzmb) and IFN-γ (Ifng) (Fig. 2, K and L), which are known to mediate tumor cell killing (26, 27). The absence of STING signaling not only abolished the dMMR-triggered inflammatory response and TIL recruitment but also enabled dMMR tumors (MLH1/STING−/− or MSH2/STING−/−) to grow comparably to WT tumors.
Because type I IFNs are key effectors of STING signaling, we next assessed the role of type I IFN signaling in dMMR tumors by treating animals with blocking anti-IFNAR1 antibodies. We observed that dMMR tumors grew more aggressively in mice treated with anti-IFNAR1, which demonstrates an essential role for type I IFN signaling in the control of dMMR tumors (fig. S3, G and H). Furthermore, we treated animals harboring MLH1- or MSH2-deficient tumors with anti-CXCR3 antibodies. These antibodies block the CXCL10-CXCR3 interaction, which is known to mediate TIL recruitment to the TME in vivo. Treatment with anti-CXCR3 also resulted in more aggressive growth of dMMR tumors in vivo, indicating that the antitumor immune responses to dMMR tumors also involve Cxcl10 signaling via the chemokine receptor CXCR3 (fig. S3, I and J). Together, these genetic in vivo data demonstrate that tumor cell–intrinsic STING signaling creates a hot cytotoxic T and NK cell–containing TME and affects in vivo tumor cell growth via type I IFN- and CXCR3-mediated mechanisms.
STINGN153S expression enforces STING signaling in cancer cells
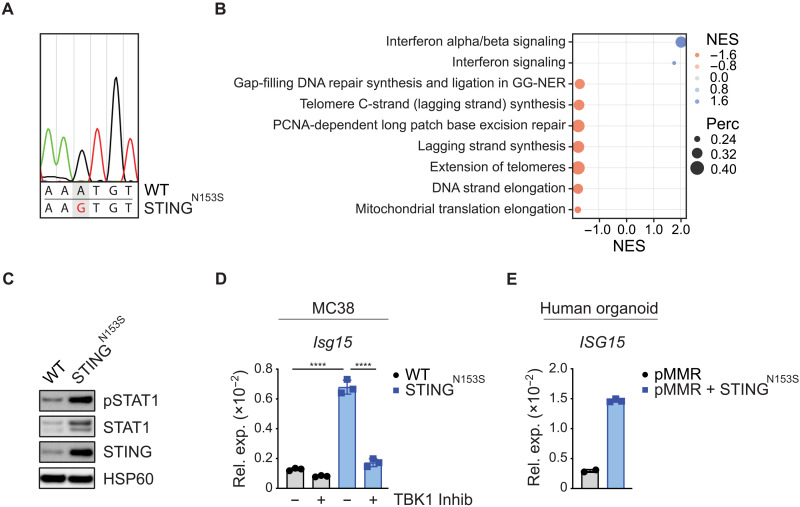
After identifying a necessity for tumor cell–intrinsic STING signaling for the immunogenicity of dMMR cancer cells, we explored whether synthetically enforced STING signaling in tumor cells could be sufficient to create hot TMEs in originally cold pMMR cancer tissues. As a strategy to genetically enforce STING signaling in MC38 cells, we used dominant active STING variants (STINGN153S), which were originally isolated from patients suffering from the inherited autoinflammatory disease STING-associated vasculopathy with onset in infancy, termed SAVI (28, 29). Using retroviral particles, we transduced STINGN153S into pMMR MC38 cells (Fig. 3A). The parental pMMR MC38 control cells are hereafter termed WT MC38 cells. Gene expression analysis and GSEA demonstrated that the expression of STINGN153S was sufficient to enrich the IFN-α/β signaling and IFN signaling gene expression signatures in pMMR MC38 cells (Fig. 3B). Moreover, expression of STINGN153S resulted in increased IFN-β production (fig. S4A). In line with the increased IFN-β production, STINGN153S induced STAT1 phosphorylation (Fig. 3C) and increased STAT1 protein expression, which is known to be induced by IFN signaling in a feed-forward loop (30), indicating constitutive signaling. Furthermore, expression of STINGN153S triggered ISG expression in a TBK1-dependent manner (Fig. 3D). In human pMMR CRC organoids, the ectopic expression of STINGN153S was also sufficient to activate the IFN pathway (Fig. 3E).
Fig. 3. A strategy to genetically enforce STING signaling in cancer cells.
(A) Electropherogram displaying the sequencing result of the PCR-amplified transgene STINGN153S from genomic DNA of MC38 tumor cells. (B) GSEA of RNA-seq data to identify differentially expressed gene sets between WT versus STINGN153S MC38 tumor cells by using the Reactome database with a percentage cutoff of >0.2 (+ve NES: STINGN153S). (C) Phosphorylation of STAT1 in cultured MC38 cells was detected by Western blotting. (D) The relative gene expression of Isg15 in WT versus STINGN153S MC38 tumor cells 16 hours after TBK1 inhibitor treatment was quantified by qPCR. (E) The relative gene expression of ISG15 in pMMR (PDO#12) and STINGN153S-transduced (=pMMR+STINGN153S) cultured primary organoids was quantified by qPCR. The data represent n = 3 independent experiments (D) or n = 2 to 3 technical replicates (E). One-way ANOVA (D) or Student’s t test (E) was used to determine significance.
Upon activation, STING requires the phosphorylation of a specific serine at position 366 (S365 in mouse STING) to drive the transcription of type I IFN genes (31). To test whether the observed effects were specifically triggered by the STINGN153S mutation, we next generated MC38 tumor cells that express the selective IFN-inactive STING mutant S365A in combination with the N153S mutant (STINGN153S/S365A) or the WT variant (STINGWT). IFN-β production (fig. S4A), STAT1 phosphorylation (fig. S4B), and Isg15 expression (fig. S4C) triggered by the constitutively active STINGN153S mutant were completely abrogated in MC38 tumor cells that expressed the double-mutant STINGN153S/S365A and were also not induced by the STINGWT variant. Thus, even in the presence of an intact MMR response, the engineered STINGN153S constructs specifically drove cell-autonomous IFN signaling in both murine and human colon cancer models.
Synthetically enforced STING signaling promotes antitumor immunity
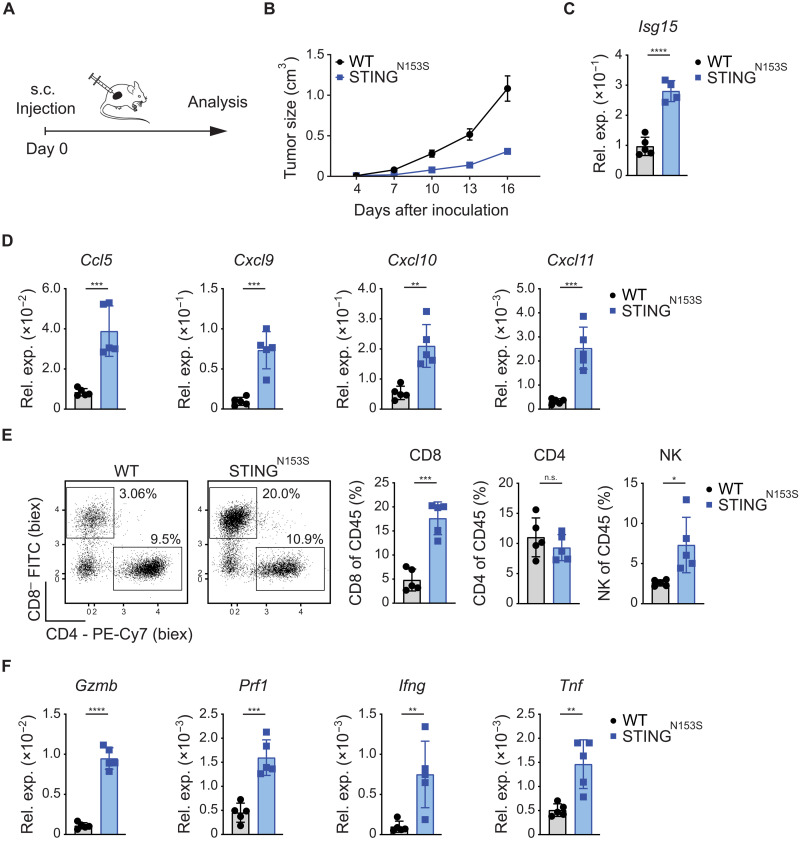
To explore the capacity of STINGN153S to induce immunogenic TMEs in originally cold tumors, we next tested the effects of tumor cell–intrinsic STINGN153S signaling in immunocompetent animals. To this end, we subcutaneously injected STINGN153S-transduced MC38 or parental WT MC38 cells into syngeneic mice and first monitored tumor growth (Fig. 4A). Similar to MMR deficiency (see Fig. 2, E and F), enforced STINGN153S signaling in MC38 cells significantly reduced tumor growth in vivo (Fig. 4B and fig. S5A), although the proliferation of STINGN153S-transduced MC38 cells in vitro was similar to that of their WT counterparts (fig. S5B). Next, we injected STINGN153S-transduced MC38 cells into immunocompromised nonobese diabetic–severe combined immunodeficient (NOD-SCID) mice that lack an intact lymphoid compartment (32). In these animals, the STINGN153S-expressing MC38 tumors grew similarly to WT MC38 tumors (fig. S5C), indicating that the control of STINGN153S-transduced MC38 cells in WT mice depends on the activation of lymphocytes. To test whether this immune-mediated growth inhibition of STINGN153S-expressing tumors was specifically dependent on STINGN153S-triggered IFN signaling, we also injected STINGN153S/S365A- and STINGWT-transduced MC38 control cells into syngeneic mice. In contrast to STINGN153S-expressing tumors, STINGN153S/S365A tumors grew similarly to STINGWT tumors, demonstrating that active STINGN153S signaling is responsible for growth inhibition in vivo (fig. S5, D and E).
Fig. 4. Synthetically enforced STING signaling promotes antitumor immunity.
(A) Schematic representation of the experimental setup in vivo. (B) Growth of subcutaneously inoculated WT and STINGN153S MC38 tumor cells in syngeneic WT C57Bl/6 mice (n = 5). (C to F) Subcutaneously grown tumors were explanted on day 17 for qPCR and FACS analysis. (C) The relative gene expression of (C) ISGs (Isg15) and (D) chemokines (Ccl5, Cxcl9, Cxcl10, Cxcl11) was quantified by qPCR. (E) FACS results. Representative dot plots and percentages (CD4, CD8, NK1.1) of live/CD45+ cells are displayed. (F) The relative gene expression of cytotoxic effector molecules (Gzmb, Prf1, Ifng, Tnf) was quantified by qPCR. The data are presented as the mean ± SEM (B). Student’s t test was used to determine significance.
Motivated by these results, we next studied the effects of tumor cell–intrinsic STINGN153S signaling on the TME in WT animals using flow cytometry and gene expression analysis. Consistent with enforced IFN signaling in STINGN153S-transduced MC38 cells in vitro, we detected strong induction of the ISG marker Isg15 in the STINGN153S-transduced tumors in vivo (Fig. 4C). Furthermore, the chemokines Ccl5, Cxcl9, Cxcl10, and Cxcl11, which mediate CTL and NK cell recruitment (25, 33, 34) into the tumor tissue, were strongly up-regulated in the TMEs of STINGN153S-expressing tumor cells (Fig. 4D). Consistently, STINGN153S-expressing tumors contained increased frequencies of CTLs and NK cells, while the frequencies of CD4+ T cells were unaltered (Fig. 4E). These infiltrating CTLs in the STINGN153S-expressing tumors were characterized by enhanced PD1 expression and increased Ifng production compared to those of tumors formed by WT MC38 cells, demonstrating that they are not only recruited to the TME but also activated in response to tumor cell–intrinsic STINGN153S signaling (fig. S5F). In line with these findings, the expression of additional cytotoxic effector molecules, such as Gzmb, perforin 1 (Prf1), Ifng, and tumor necrosis factor (Tnf), in the TME was also strongly up-regulated (Fig. 4F). Thus, genetically enforced STINGN153S signaling in pMMR tumor cells is by itself sufficient to shape an immune cell–infiltrated and immunologically active TME that exhibits the key requirements for productive antitumor immunity.
Tumor cell–intrinsic STING enforcement sensitizes ICI therapy
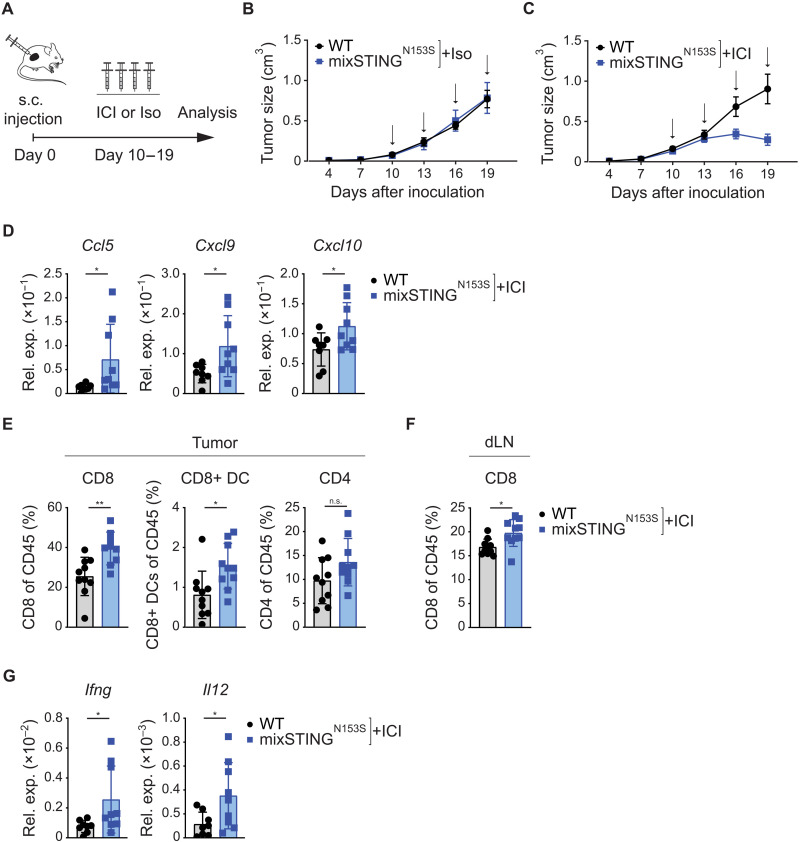
To test whether the expression of STINGN153S in a subset of cancer cells could be sufficient to sensitize the TME to ICI treatment, we mixed WT and STINGN153S-expressing MC38 cells before subcutaneous inoculation (Fig. 5A). Without additional treatment, tumors that contained less than one-third STINGN153S-expressing MC38 cells (=mixSTINGN153S) grew equally to pure WT MC38 (=WT) tumors (Fig. 5B). However, ICI treatment with anti-PD1 and anti-CTLA4 inhibitors significantly strengthened the regression of mixSTINGN153S tumors and improved survival of the animals (Fig. 5C and fig. S6B). Furthermore, within the responsive mixSTINGN153S-containing tumor tissues, we detected an enhanced inflammatory state with increased expression of Ccl5, Cxcl9, and Cxcl10 (Fig. 5D); increased frequencies of CD8+ dendritic cells (DCs) (CD11c+, CD11b−, and CD8+), which are critical for tumor antigen cross-presentation and priming of CD8+ T cells against tumor antigens (35, 36); and increases in CTL infiltration. However, there were no differences in CD4+ T cell infiltration (Fig. 5E). Increases in CTLs were also observed in the draining lymph nodes (Fig. 5F). Consistent with productive DC and cytotoxic activity, the expression of Il12, a DC-derived cytokine that shapes antitumor immunity, and Ifng was also significantly up-regulated in STINGN153S-containing tumors (Fig. 5G). Thus, genetically enforced STINGN153S signaling in a subset of tumor cells is sufficient to enhance ICI therapy responsiveness.
Fig. 5. Tumor cell–intrinsic STING enforcement sensitizes to immune checkpoint inhibitor therapy.
(A) Schematic representation of the experimental setup in vivo. Growth of subcutaneously inoculated WT or mixSTINGN153S MC38 tumors in syngeneic C57Bl/6 mice (B) treated with isotype control (=iso) (n = 4) or (C) treated with anti-PD1/anti-CTLA4 (=ICI) (n = 8 to 10) every 3 days (black arrows). (D to G) Subcutaneously grown tumors that were treated with ICI therapy were explanted on day 21 for qPCR and FACS analysis. The relative gene expression of (D) chemokines (Ccl5, Cxcl9, Cxcl10) was quantified by qPCR. FACS was performed, and the (E) percentages (CD8, CD8+ DCs, CD4) of live/CD45+ cells in the tumor and (F) percentages (CD8) of live/CD45+ cells in the draining lymph node (dLN) are shown. (G) The relative gene expression of cytokines (Ifng, Il12) was quantified by qPCR. The data are presented as the mean ± SEM (B and C). Student’s t test (D to G) was used to determine significance.
Expression of STINGN153S in a subset of tumor cells reprograms the TME
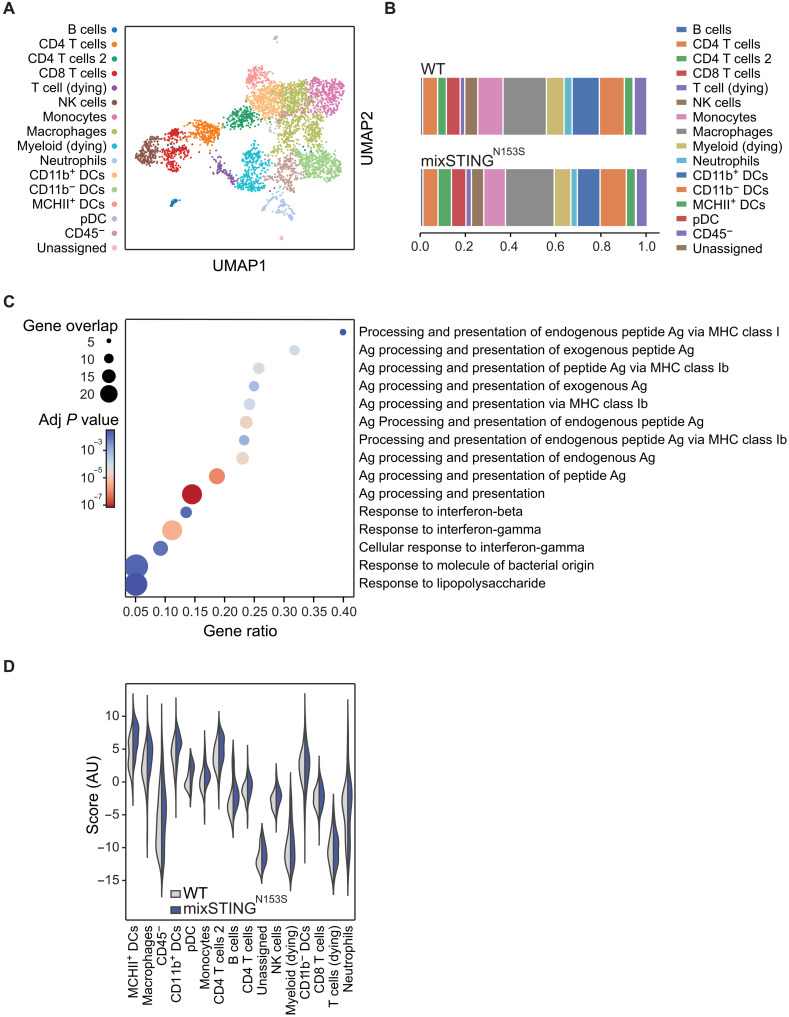
To dissect the mechanisms by which synthetically enforced STING signaling sensitizes cells to ICI treatment, we analyzed the TMEs of mixed STINGN153S tumors and WT tumors under equal growth conditions (see Fig. 5B) using cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) (37). CITE-seq is a single-cell methodology that enables transcriptomic analysis of individual cells within complex tissues that are pretagged with barcoded antibodies directed against cellular markers. After labeling the cellular suspensions from growing tumors ex vivo with a custom-generated antibody panel, we performed an integrated TME analysis on a joint embedding computed by the variational autoencoder TotalVI (38). By clustering the cells in latent space using modularity maximization, we identified 16 cell population clusters in both TMEs based on gene expression and antibody-derived tag (ADT) abundance (Fig. 6A and fig. S6A). When we compared the TMEs of mixed STINGN153S MC38 tumors to those of pure WT control MC38 tumors, we did not identify any overt differences in the overall composition of either the CD45+ immune or CD45− nonimmune cellular subpopulations (Fig. 6B and fig. S6B). This finding was further confirmed by fluorescence-activated cell sorting (FACS) analysis and cellular staining of the TME for CD4 T cells (CD4+), CD8 T cells (CD8+), NK cells (NK1.1+), DCs (CD11c+), or CD8+ DCs (CD11c+, CD11b−, CD8+) (fig. S6C). However, differential gene expression analysis within the tumor-infiltrating cells using diffxpy coupled with gene set enrichment using g:profiler on the GO:BP database revealed, specifically in the STINGN153S-containing tumors, enrichment of key antigen processing and presentation pathways such as “processing and presentation of endogenous peptide antigens via major histocompatibility complex class I (MHCI),” as well as IFN signaling signatures such as the “response to IFN-β” and “response to IFN-γ” (Fig. 6C). Next, we applied an eigengene score approach to identify the cellular clusters that contribute to these overall differential expression signatures (Fig. 6D). Several types of APCs, including macrophages and DC subsets, such as MHCII+ DCs, CD11b+ DCs, and plasmacytoid DCs (pDCs), together with the CD45− nonimmune cells, were among the strongest contributors to the enhanced activation of antigen presentation and IFN signaling in the TMEs of STINGN153S-expressing tumors (Fig. 6D). Among these, a strong enrichment for antigen processing and presentation as well as IFN signaling was particularly detected in the two DC clusters “MHCII+ DCs” and “CD11b+ DCs” (fig. S6D). These cells represent APC populations that are important for the initiation of antitumor immunity. Thus, synthetic enforcement of STINGN153S signaling in a fraction of tumor cells is sufficient to reprogram the TME of originally cold tumors and sensitize these tumors to ICI therapy.
Fig. 6. The expression of STINGN153S in a subset of tumor cells reprograms the TME.
CITE-seq analyses of subcutaneously grown WT and mixed STINGN153S MC38 tumors. (A) UMAP plot of annotated clusters displaying the individual clusters. (B) Cellular composition of cell clusters in WT and mixed STINGN153S tumors. (C) Differentially expressed gene sets determined by GSEA by using g:profiler for all GO:BP terms enriched for fewer than 400 genes considering all clusters. (D) Violin plot displaying how strongly the different clusters contribute to the differentially expressed genes observed in (C).
DISCUSSION
By exploring mechanisms that promote antitumor immunity in dMMR CRC, we found that defects in the MMR machinery not only increase TMB but also result in an accumulation of DNA in the cancer cell cytosol, which stimulates the innate immune sensor cGAS to activate STING signaling. This tumor cell–intrinsic pathway is key to shaping a hot TME with productive antitumor activity. These findings are in line with two independent studies that recently also reported that MLH1-mutated CRC cells engage STING signaling for antitumor immunity (19, 39). We additionally demonstrate that MSH2 deficiency activates the cGAS-STING-IFN pathway and provide data from dMMR and pMMR CRC patients and patient-derived CRC organoids, demonstrating that dMMR triggers epithelial cell–intrinsic IFN engagement in human CRC. Thus, the loss of genomic integrity in CRC cells induces tumor cell–intrinsic STING and IFN activation and an antitumorigenic inflammatory TME.
On the basis of these insights, we envisioned that synthetically enforced STING signaling within tumor cells may be sufficient to create hot TME niches in originally cold tumor tissues. To test this, we used SAVI-STING variants isolated from patients with an autoinflammatory disorder that are characterized by gain-of-function point mutations near the STING dimerization site (28). Ectopic expression of gain-of-function STINGN153S was sufficient to induce cell-autonomous STING signaling in the absence of MMR deficiency in murine and human CRC. These STINGN153S-expressing cancer cells created hot TMEs characterized by excessive expression of TIL-recruiting chemokines and strong infiltration of activated CTLs and NK cells with expression of cytotoxic effector molecules such as Gzmb, Prf1, Ifng, and Tnf. Thus, synthetically enforcing STING signaling in cancer cells is sufficient to induce those inflammatory cues that are needed to recruit and prime CTLs into the TME even in the absence of a hypermutator phenotype.
Single-cell analysis demonstrated that the introduction of constitutively active STING into only a subset of cancer cells already induces inflammatory remodeling of the TME with changes in gene expression signatures within defined immune cell subpopulations that indicate up-regulated APC functionality and antigen processing in DCs, which are critical for tumor antigen cross-presentation and coupling of innate to adaptive immunity. Consistently, tumors that contained STINGN153S-expressing cancer cells were sensitized to ICI therapy with superior antitumor immune responses upon anti-PD1 and anti-CTLA4 treatment. Because effective ICI responses require a productive DC:T cell interaction (40), our data provide proof of principle that synthetically enforced STING signaling in tumor cells promotes APC:CTL cross-talk in the TME via activation of APCs, production of chemokines and cytokines that attract TILs, and priming of CTLs and indicate that this strategy could be further explored as a therapeutic concept to sensitize tumor tissues to ICIs. While our study was triggered by the observation that STING signaling in dMMR CRC shapes effective antitumor immunity, it is conceivable that genetically enforced STINGN153S signaling could also sensitize cancer tissues beyond CRC, which needs to be further investigated.
Our strategy of TME reprogramming via synthetically enforced STING signaling in cancer cells could offer some advantages over current TME-modulating approaches that inject small-molecule STING agonists in vivo (41–43). On the one hand, small-molecule STING activators exert unwanted side effects on immune effector cells, including CD8+ T cells, in which STING signaling blocks proliferation and induces apoptosis; this disables CTL function (44–47) or results in the recruitment of suppressor cells and up-regulation of inhibitory molecules such as PDL1 that impede the antitumor immune response (48–50). On the other hand, endogenous STING in tumor cells is frequently down-regulated or inhibited (51–54), resulting in inadequate pharmacological targetability by small molecules. These mentioned difficulties are exemplary reasons illustrating that although there are constantly new small-molecule STING agonist formulations being developed and tested, their success is limited (42, 55). Our concept of synthetically enforcing STING signaling selectively in tumor cells could in principle overcome these hurdles. However, for clinical translation, several key points must be addressed. The most important next step is the development of effective protocols for constitutively activating STING gene transfer into the tumor cells of patients. One possibility is the isolation of cancer cells from biopsies and ex vivo manipulation and reinjection of engineered STING-expressing cells into the tumor tissues. Alternatively, gene transfer protocols based on viral vectors (56), mRNA vaccines (57, 58), or cell-directed lipid nanoparticles (59, 60) could be used to express synthetically constitutively active STING variants in tumor cells in vivo. Because selective activation of STING signaling in cancer cells is sufficient to appropriate the TME for ICI responsiveness, this approach is a promising TME-modulating strategy to combine with ICI therapy and to hopefully provide additional clinical benefits for cancer patients.
MATERIALS AND METHODS
Study design
This was a preclinical and translational study with the aim of investigating the mechanisms governing ICI therapy responsiveness in dMMR tumors and potentially using these mechanisms to sensitize pMMR tumors to ICI therapy. For TMB assessment and transcriptional analysis of human CRC, we used publicly available datasets from the TCGA and patient-derived primary organoids. To study MMR deficiency in tumor cells, we genetically modified murine MC38 tumor cells with CRISPR-Cas9 for cell culture assays in vitro and for subcutaneous tumor models in vivo. After establishing the role of STING signaling in the antitumor immunity of dMMR tumors, we tested the impact of constitutive STING activation in pMMR tumors. For this, patient-derived primary CRC organoids and murine MC38 tumor cells were retrovirally transduced with a constitutively active STING variant for cell culture assays in vitro and for subcutaneous tumor models in vivo. To test the therapeutic potential of tumor cell–intrinsic constitutively active STING signaling in combination with ICI therapy, subcutaneously grown tumors in recipient mice were treated with ICI therapy. Investigators were blinded to the tumor size measurements. The animal numbers are stated in the figure legends, and replicates are presented as individual dots. The statistical methods are described below and indicated in the figure legends.
Derivation and biobanking of CRC organoids
Resection samples from CRC patients were provided by the University Cancer Center Frankfurt (UCT). All materials were collected as part of the interdisciplinary Biobank and Database Frankfurt (iBDF) after written informed consent was obtained, and the study was approved by the institutional review board of the UCT and the Ethical Committee at the University Hospital Frankfurt (ethics vote: 4/09; project numbers: SGI-06-2015 and SGI-12-2018). After pathological assessment of the tumor and adjacent normal regions, the samples were rapidly processed on ice. Necrotic regions were removed, and tissues were cut with scalpels into pieces (~1 to 2 mm diameter). Three to five randomly chosen pieces were immediately frozen at −80°C for subsequent DNA/RNA isolation. WES and RNA-seq were performed as part of a CRC organoid-stroma biobank that will be published elsewhere (manuscript in revisions).
WES and RNA-seq of CRC organoids
For WES analysis, from each sample, genomic DNA from tumor, tumor organoids, and normal adjacent tissue was subjected to whole-exome analysis. A total of 450 ng of genomic DNA was used for library preparation using an Agilent Low Input Exome-Seq Human v6 kit. Indices were introduced using an Agilent SureSelect XT kit, and sequencing was performed on HiSeq 4000 at the Genomics and Proteomics Core Facility at the DKFZ Heidelberg. Pathological variants were detected by following GATK best practices (61) using Mutect2 (GATK3 v3.8). In addition, we used the variant callers Mutect1 v1.1.7 (62) and VarScan2 v2.4.3 (63). Only variants with a variant allele frequency (VAF) cutoff of 5% identified by Mutect2 and confirmed by one of the other two variant callers were considered and annotated using the Ensembl variant effect predictor (VEP v99) (64). Identified mutations in MMR genes are listed in table S1. Total number of detected mutations and the sum of allele frequencies of the identified variants of the indicated recurrent mutations are listed in table S2. For RNA-seq analysis, the total RNA concentration was measured using a Qubit RNA HS Assay Kit assay (Thermo Fisher Scientific). For library generation, a SMARTer Stranded Total RNA Sample Prep Kit–HI Mammalian kit (Takara Bio) was used following the manufacturer’s recommendations. One microgram of input total RNA was fragmented in Bioruptor (Diagenode) for 5 min for samples with RNA integrity numbers (RINs) of >7 and 4 min for samples with RINs of 4 to 7 and chilled on ice. The final libraries were measured via Qubit und ScreenTape analysis before multiplexing. RNA-seq was performed using a TruSeq RNA 50 cycle kit (Illumina) and run on a HiSeq 2000 instrument (Illumina) at the Genomics and Proteomics Core Facility at the DKFZ Heidelberg. After the data quality was checked using FastQC, the reads were aligned using STAR version 2.7a. The read counts were computed by the STAR function “quantMode” using the reference genome GRCh38. Gene annotation was performed using the biomaRt R package (version 2.38.0). Differential gene expression analysis was performed in an unpaired manner using the DESeq2 R package (versions and 1.32.0). GSEA was performed using the fgsea R package (version 1.18.0) as reported previously (65). Genes with a base mean < 20 were removed before they were ranked ascendingly based on the log2 fold change. The ranked gene list was then compared against the 1635 Reactome gene sets of the MSigDB database (version 7.4) using the “fgsea” function with default settings. Normalized enrichment scores (NESs) and P values were used for subsequent analyses. The differential gene expression analysis results between dMMR and pMMR organoid samples are shown in table S3. For fig. S1A, all Reactome gene sets with P ≤ 0.01 were plotted in a bubble chart, with one bubble representing one gene set. The bubble color depicts the negative decadic logarithm of the P value, while the bubble size depicts the fraction of the number of leading edge genes as reported by fgsea over the number of total genes in the gene set.
Human COADREAD samples (TCGA)
Clinical and mutation analysis data for the Colorectal Adenocarcinoma TCGA PanCancer Atlas dataset (https://pubmed.ncbi.nlm.nih.gov/29596782/) were downloaded via the cBioPortal for Cancer Genomics (https://pubmed.ncbi.nlm.nih.gov/22588877). In total, 524 tumor samples with complete clinical and mutation calling information were available (as of February 2020). Mutation calls for the four genes MLH1, MSH2, POLE, and POLD1 were used to assign each tumor sample to one of two categories: pMMR, with WT MLH1, MSH2, POLE, and POLD1, and dMMR, with at least one mutation in MLH1, MSH2, POLE, or POLD1. The mutations considered were missense, nonsense (premature stop codon), frameshift insertion or deletion, in-frame deletion, and splice-site mutations (see table S1). The tumor sample mutation counts reported by TCGA were normalized to a human whole-exome size of 30 Megabase pairs (Mbp) and reported as the mutation count per Mbp. For the same 524 tumor samples, the gene expression values for 20,531 genes obtained by RNA-seq were downloaded via cBioPortal. The siggenes R package (version 1.72.0) was used to identify genes differentially expressed between dMMR and pMMR tumors. The differentially expressed genes were ranked by their fold change, and the ranked gene list was used as input for a preranked GSEA using the fgsea R package (version 1.18.0) against the Reactome gene sets of the MSigDB database (version 7.4) as available in the msigdbr R package (version 7.5.1). For Fig. 1B, all Reactome gene sets with an adjusted P ≤ 0.05 were plotted in a bubble chart, with one bubble representing one gene set. The bubble color depicts the NES, while the bubble size depicts the fraction of the number of leading edge genes as reported by fgsea over the number of total genes in the gene set.
Mice
All animal work was conducted in accordance with German Federal Animal Protection Laws and approved by the government of Upper Bavaria (Regierung von Oberbayern, Munich, Germany). For murine tumor transplantation experiments, C57BL/6 mice were purchased from Charles River Laboratories, and NOD-SCID mice were purchased from The Jackson Laboratory (stock no. 001303). Female 6- to 8-week-old mice were used for experiments. Tumor size was measured with an electronic caliper every 3 days and calculated by the following formula: V = (W2 × L)/2, where W is the width (minor tumor axis) and L is the length (major tumor axis). Mice were assessed by their general behavior, outer appearance, body condition parameters, and body weight.
Cell culture
The mouse colon adenocarcinoma cell line MC38 was donated by B. Nordic and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco: 41966-029) containing 10% fetal calf serum (FCS), 1% penicillin/streptomycin, 1% sodium pyruvate, 1% nonessential amino acids, 2% Hepes, and 0.05 mM β-mercaptoethanol. Phoenix-Eco cells and human embryonic kidney (HEK)–293T cells were cultured in DMEM (Gibco: 41966-029) containing 10% FCS, 1% penicillin/streptomycin, 1% nonessential amino acids, and 1% sodium pyruvate. Organoid lines were maintained in complete tumor expansion medium as described previously bySchnalzger et al. (67). All cells were cultured under standard cell culture conditions at 37°C in 5% CO2 and 95% humidity. The cells were routinely tested for mycoplasma contamination.
Tumor experiments and treatments
MC38 tumor cells [0.5 × 106, mixed 1:1 in phosphate-buffered saline (PBS):Matrigel (Corning)] were injected subcutaneously into the flanks of the recipient mice. Tumor-bearing mice were treated with 250 μg of anti-PD1 (clone RPM1-14)/200 μg of anti-CTLA4 (clone 9H10) (both Bio X Cell) or equal quantities of the respective isotype controls rat immunoglobulin G2a (IgG2a) (clone 2A3)/polyclonal Syrian hamster IgG (both from Bio X Cell) by intraperitoneal administration. The mice were treated every 3 days. For anti-IFNAR1 and anti-CXCR3 inhibitor experiments, tumor-bearing mice were treated with or without anti-IFNAR1 (200 μg per mouse) (clone: MAR1-5A3, Bio X Cell) or anti-CXCR3 (200 μg per mouse) (clone: CXCR3-173; Bio X Cell) at days 0, 3, 6, 9, 12, and 15 after inoculation.
Gene editing
To generate knockout cell lines, single-guide RNA (sgRNA) sequences were designed using the CRISPR tool (http://crispr.mit.edu), cloned into the CRISPR-Cas9 system plasmid pSpCas9(BB)-2A-Puro (PX459) (a gift from F. Zhang, Addgene, plasmid no. 62988) or pSpCas9(BB)-2A-Neo (a gift from K.-I. Takemaru, Addgene, plasmid no. 127762), and transfected into MC38 tumor cells with Lipofectamine 3000 (Thermo Fisher Scientific). After 24 hours, the transfected cells were selected with the appropriate antibiotics for 3 to 5 days and subsequently seeded in 96-well plates at one cell per well. Expanded cells were then harvested, and knockout was validated by Western blotting. sgRNA sequence: MLH1 sgRNA, GATGGTCCGTACGGACTCCC; MSH2 sgRNA, AACATACATTCGTCAGACCG; cGAS sgRNA, CGAGGCGCGGAAAGTCGTAA; STING sgRNA, GTACCCAATGTAGTATGACC.
Retroviral modification of murine tumor cells and human organoids
Mouse STINGN153S, STINGN153S/S365A, and STINGWT variants were cloned into the MSCV-puro vector (a gift from T. Jacks, Addgene, plasmid no. 68469) and the pMSCV-blasticidin vector (a gift from D. Mu, Addgene, plasmid no. 75085) using standard cloning techniques. MC38 tumor cells were transduced with STING variant–containing vectors. Retroviral particles were produced with the Phoenix-Eco packaging cell line. Supernatants were collected 48 hours after transfection, filtered through a 0.45-μm filter, and used fresh or frozen at −80°C. Cells were infected with the virus in the presence of protaminsulfat (Sigma-Aldrich). Thereafter, cells with stable expression were selected by the addition of puromycin (5 μg/ml; InvivoGen) or blasticidin (25 μg/ml; InvivoGen) to the culture medium. The organoid lines were transduced with MSCV-puro-STINGN153S or as a control with pMSCV-FLIP-puro-dsRed-GFP-miRNA (a gift from B.-K. Koo, Addgene, plasmid no. 32704). Retroviral vectors were packaged in HEK-293T cells using pcDNA3.MLVgp (66) and pCMV-VSV-G (a gift from B. Weinberg, Addgene, plasmid no. 8454) following the procedure described by Schnalzger et al. (67). Cells with stable expression were selected by the addition of puromycin (1 μg/ml) to the culture medium for two passages.
In vitro proliferation assay
MC38 tumor cells (5 × 103) were seeded in a flat-bottom 96-well plate. At each time point, absorbance was measured using the CellTiter96 AQueous One Solution Cell Proliferation Assay Kit (Promega) according to the manufacturer’s instructions.
Quantitative polymerase chain reaction
RNA was isolated from 105 MC38 tumor cells 24 hours after seeding using the RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. For organoid lines, organoids were seeded in triplicate wells and cultured for 3 days in tumor expansion medium without puromycin before a medium change and an additional 16 hours of culture. Total RNA was collected using a NucleoSpin-RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. Tumor tissue was homogenized in gentleMACS M Tubes (Miltenyi) using the gentleMACS Dissociator (Miltenyi), and then RNA was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. The concentration of RNA was measured with a NanoDrop. RNA was then reverse-transcribed into complementary DNA (cDNA) using qScript reagent (Quantabio), and real-time polymerase chain reaction (PCR) was performed using Takyon No ROX SYBR mix (Eurogentec). The gene expression levels were calculated by the ∆∆Ct method and normalized to those of Gapdh.
The following murine primer sequences were used: Gapdh, 5′-AACAGCAACTCCCACTCTTC-3′ (forward) and 5′-CCTGTTGCTGTAGCCGTATT-3′ (reverse); Isg15, 5′-GGTGTCCGTGACTAACTCCAT-3′ (forward) and 5′-CTGTACCACTAGCATCACTGTG-3′ (reverse); Ccl5, 5′-GCTGCTTTGCCTACCTCTCC-3′ (forward) and 5′-TCGAGTGACAAACACGACTGC-3′ (reverse); Cxcl9, 5′-TCCTTTTGGGCATCATCTTCC-3′ (forward) and 5′-TTTGTAGTGGATCGTGCCTCG-3′ (reverse); Cxcl10, 5′-CCAAGTGCTGCCGTCATTTTC-3′ (forward) and 5′-GGCTCGCAGGGATGATTTCAA-3′ (reverse); Cxcl11, 5′-GGCTTCCTTATGTTCAAACAGGG-3′ (forward) and 5′-GCCGTTACTCGGGTAAATTACA-3′ (reverse); Gzmb, 5′-CCACTCTCGACCCTACATGG-3′ (forward) and 5′-GGCCCCCAAAGTGACATTTATT-3′ (reverse); Ifng, 5′-CAGCTCCAAGAAAGGACGAAC-3′ (forward) and 5′-GGCAGTGTAACTCTTCTGCAT-3′ (reverse); Prf1, 5′-AGCACAAGTTCGTGCCAGG-3′ (forward) and 5′-GCGTCTCTCATTAGGGAGTTTTT-3′ (reverse); Tnf , 5′-ATGAGCACAGAAAGCATGATC-3′ (forward) and 5′-TACAGGCTTGTCACTCGAATT-3′ (reverse); Il12, 5′-TGGTTTGCCATCGTTTTGCTG-3′ (forward) and 5′-ACAGGTGAGGTTCACTGTTTCT-3′ (reverse). The following murine primer sequences for cytosolic DNA quantification were used: Gapdh, 5′-CAACTGCTTAGCCCCCCTGG-3′ (forward) and 5′-GCAGGGTAAGATAAGAAATG-3′ (reverse). The following human primer sequences were used: GAPDH, 5′-AGCCACATCGCTCAGACAC-3′ (forward) and 5′-GCCCAATACGACCAAATCC-3′ (reverse); ISG15, 5′-GCGAACTCATCTTTGCCAGTA-3′ (forward) and 5′-CCAGCATCTTCACCGTCAG-3′ (reverse).
Inhibitor treatment
A total of 105 MC38 cells were seeded in a flat-bottom 96-well plate. The cells were treated with 10 μM TBK1 inhibitor (InvivoGen) or control medium for 16 hours. Total RNA was collected using the RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. Organoid fragments were generated by dissociation and seeding in Growth Factor Reduced Matrigel (Corning) and cultured for 3 days before medium change and addition of 10 μM TBK1 inhibitor or control complete tumor expansion medium for an additional 16 hours. The experiments were performed in triplicate wells, and RNA was collected using a NucleoSpin-RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. For anti-IFNAR1 inhibitor experiments, RNA was isolated from MC38 tumor cells 24 hours after treatment with anti-IFNAR1 (30 μg/ml) (clone: MAR1-5A3; Bio X Cell) by using the RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed as described in the previous section.
Isolation of cytosolic DNA
After 48 hours of culture, the cytosolic fraction of 5 × 106 MC38 tumor cells was isolated using a mitochondrial isolation kit (Thermo Fisher Scientific) as reported (19). The manufacturer’s protocol was followed until the cytosolic fraction was obtained. Then, DNA from the cytosolic fraction was isolated using a QIAGEN DNeasy blood and tissue kit (QIAGEN) according to the manufacturer’s instructions. The DNA amount was measured with Qubit 4 using the Qubit 1× dsDNA HS Assay Kit following the manufacturer’s instructions. To quantify the relative amount of DNA in the cytosolic fraction, qPCR with primers specific for genomic DNA was performed as described in the “Quantitative polymerase chain reaction” section. The relative DNA amount was calculated by normalization to the DNA amount of the WT condition.
Enzyme-linked immunosorbent assay
After 24 hours of culture, 2.5 × 105 MC38 tumor cells were washed with ice-cold PBS and harvested in M-PER Buffer (Thermo Fisher Scientific). The lysates were clarified by centrifugation at 20,000g/4°C for 10 min and then stored at −80°C or used fresh. cGAMP enzyme-linked immunosorbent assay (ELISA) was then performed according to the manufacturer’s instructions (Cayman Chemical). To measure IFN-β in the cell culture medium, MC38 tumor cells were cultured for 48 hours, and the supernatants were harvested and stored at −80°C or used fresh. Then, mouse IFN-β ELISA was performed according to the manufacturer’s instructions (PBL Assay Science).
Immunoblotting
MC38 tumor cells were washed with ice-cold PBS and harvested in radioimmunoprecipitation assay buffer (Sigma-Aldrich) supplemented with protease inhibitors, 10 mM NaF, and 4 mM Na3VO4 (Calbiochem). The lysates were clarified by centrifugation at 20,000g at 4°C, and the protein concentration was determined with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). The proteins in 15 μg of sample were denatured with NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) for 10 min at 70°C, subsequently separated on a 10% polyacrylamide gel, transferred onto a nitrocellulose membrane (Cytiva), blocked with 5% nonfat dry milk in tris-buffered saline with Tween 20 (TBST) buffer (0.1% Tween 20) for 1 hour, and probed with the following primary and secondary antibodies: MLH1 (ab92312, Abcam), MSH2 (ab70270, Abcam), cGAS (#31659, Cell Signaling Technology), STING (#13647, Cell Signaling Technology), STING (#50494, Cell Signaling Technology), phospho-STAT1 (#8826, Cell Signaling Technology), phospho-STAT1 (#9167, Cell Signaling Technology), STAT1 (#9172, Cell Signaling Technology), HSP60 (BD Biosciences), anti-mouse IgG–horseradish peroxidase (HRP) (Cell Signaling Technology), and anti-rabbit IgG-HRP (Cell Signaling Technology). Visualization was performed by using Pierce ECL Western blotting substrates (Thermo Fisher Scientific).
Sample preparation—flow cytometry
For FACS analysis, MC38 mouse tumors were cut into small pieces, dissociated using a mouse tumor dissociation kit (Miltenyi) with the gentleMACS Octo Dissociator (Miltenyi), and filtered through 100- and 30-μm strainers. Then, immune cells were isolated from the resulting single-cell suspension using mouse CD45 (TIL) MicroBeads (Miltenyi) with a MACS Separator (Miltenyi). For intracellular cytokine staining, cells were incubated with 100 nM phorbol 12-myristate 13-acetate, 1 μM ionomycin (both Sigma-Aldrich), and brefeldin A (BioLegend) for 4 hours at 37°C. The cells were then stained with Fixable Viability Dye (eBioscience). After blocking with anti-CD16/32 and anti-CD16.2 (both from BioLegend), the following fluorochrome-coupled antibodies were used for flow cytometric analysis:
| Marker | Fluorophore | Clone |
| CD45 | PerCP-Cy5.5 | 30F-11 |
| TCRb | APC-Cy7 | H57-597 |
| CD4 | PE-Cy7 | GK1.5 |
| CD8 | FITC | 53-6.7 |
| IFNg | PE | XMG1.2 |
| PD1 | BV421 – ef450 | 29F.1A12 |
| NK1.1 | APC | PK136 |
| CD11c | BV785 | N418 |
| CD11b | AF700 | M1/70 |
| CD19 | BUV395 | 1D3 |
Data were collected with LSRFortessa (BD Biosciences) and analyzed using FlowJo (BD Biosciences).
RNA sequencing
After 24 hours of culture, RNA was isolated from 105 MC38 tumor cells using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Next, library preparation for bulk sequencing of poly(A)-RNA was performed as described previously (68). Briefly, the barcoded cDNA of each sample was generated with Maxima RT polymerase (Thermo Fisher Scientific) using oligo-dT primers containing barcodes, unique molecular identifiers (UMIs), and an adaptor. The ends of the cDNAs were extended by a template switch oligo (TSO), and full-length cDNA was amplified with primers binding to the TSO site and the adaptor. An NEB UltraII FS kit was used to fragment cDNA. After end repair and A-tailing, a TruSeq adapter was ligated, and 3′-end fragments were finally amplified using primers with Illumina P5 and P7 overhangs. In comparison to the method of Parekh et al. (68), the P5 and P7 sites were exchanged to allow sequencing of the cDNA in read1 and barcodes and UMIs in read2 to achieve better cluster recognition. The library was sequenced on a NextSeq 500 (Illumina) sequencer with 63 cycles for the cDNA in read1 and 16 cycles for the barcodes and UMIs in read2. The data were processed using the published Drop-seq pipeline (v1.0) to generate sample- and genewise UMI tables (69). The reference genome (GRCm38) was used for alignment. Transcript and gene definitions were used according to GENCODE version M25. Differential gene expression was assessed with the DESeq2 package (70) in R (version 3.6.2). Preranked GSEA was performed with the gseapy Python package (version 0.10.1) (71) using the Reactome pathway database.
Cellular indexing of transcriptomes and epitopes by sequencing
Sample and library preparation
For both the pure WT MC38 and mixed STINGN153S conditions, three subcutaneously grown tumors (see the “Tumor experiments and treatments” section) were harvested, and the isolated cells were pooled into one sample; labeled with CITE-seq antibodies against CD45 (clone 30-F11), CD3 (clone 17A2), CD4 (clone RM4-5), CD8a (clone 53-6.7), CD11b (clone M1/70), CD11c (clone N418), NK1.1 (clone PK136), CD19 (clone 6D5), Ly6C (clone HK1.4), Ly6G (clone 1A8), F4/80 (clone BM8), I-A/E-I (MHCII) (clone M5/114.15.2), CD279 (PD1) (clone RMP1-30), and CD274 (PDL1) (clone MIH6); and enriched for live/CD45+ cells by sorting (BD FACSAria Fusion). For each genotype, CD45+ and CD45− cells were pooled in a 1:1 ratio. The sorted cells were then washed once with PBS + 2% FCS and subsequently counted to determine the exact cell number. The fraction of dead cells was estimated by trypan blue staining. The pooled cell suspensions were immediately used for single-cell RNA-seq with feature barcoding library preparation with a target recovery of 10,000 cells. Libraries were prepared using Chromium Single Cell 3′ Reagent Kit v3.1 (10X Genomics, PN-1000269) and the 3′ Feature Barcode Kit (10X Genomics, PN-1000262) according to the manufacturer’s instructions. All libraries from gene expression and feature barcoding were pooled and sequenced according to 10X Genomics’ recommendations on an Illumina NovaSeq 6000 system with a target read depth of 50,000 reads/cell gene expression libraries and 5000 reads/cell for feature barcoding libraries.
Raw data processing
The raw read data were mapped to version GRCm38 release 101 with Cell Ranger.
Quality control and preprocessing
Count data tables were loaded into and analyzed in Scanpy (version 1.7.2) (72) according to a recently published best-practices pipeline (73). Quality control of the mapped data was performed separately for ADT and for RNA data based on the joint distribution of count depth and the number of genes expressed. The ADT data were filtered to a minimum of 800 and a maximum of 15,000 counts, and the RNA data were filtered to a minimum of 200 and a maximum of 10,000 genes. Only cells passing both thresholds were retained for downstream analysis, leaving a dataset of 4086 cells that passed this filtering. To make the cellular profiles comparable and remove the effects of sequencing depth, we normalized the RNA and ADT data. The RNA data were normalized using the scran pooling method (74) via the computeSumFactors() function implemented in the Scran package (version 1.14.1) and subsequently log+1-transformed. ADT data were normalized using the Seurat implementation (version 3.0.2) (75) of the centered log ratio transform, clr
where g(x) is the geometric mean of the vector, x represents the cells, and i to D represent all ADT features.
Clustering and annotation
To generate joint embedding of the protein and the RNA data, TotalVI (as implemented in scvi-tools version 0.13.1) (38) was used with the parameters n_layers=2 and latent_distribution=‘normal’. A k-nearest neighbor graph was computed on the latent space generated by TotalVI using the Euclidean norm to compute the k = 15 nearest neighbors via the Scanpy function sc.pp.neighbors. To visualize the data, a Uniform Manifold Approximation and Projection (UMAP) (76) representation (package: umap-learn, version 0.5.1) was computed on this neighborhood graph. Cells were clustered with the Leiden algorithm (package: leidenalg, version 0.8.7) (77) on that neighborhood graph using resolution = 1. The clusters were then annotated jointly using the protein and RNA data. To make the results of this analysis comparable to the FACS data, the clusters were annotated predominantly on the basis of the protein abundance levels.
Differential expression analysis
Because of the small number of cells per cell type and the small total expression changes, we first tested differential expression over all cell types. Differential expression between the WT and the STINGN153S mutant was tested with Diffxpy (version 0.7.4+18.gb8c6ae0) (78). Diffxpy fits a negative binomial model to the raw count data and allows the addition of covariates into the model. Here, we fit the model
where the condition is either WT STING or the STINGN153S mutant in a one-hot encoded covariate. Furthermore, we add the size factors from Scran as an offset to the model. The differential test was performed via a Wald test over the condition covariate per gene for all genes expressed in at least 50 cells in the tested cluster. Multiple testing correction was performed via the Benjamini-Hochberg method (79). Differentially expressed genes were filtered to a corrected P value below 0.05.
Gene set scoring
As described, per cell type, differential expression tests did not have enough power. To still identify the population from which the differential expression signature arose, we used singular value decomposition–based scoring by performing principal components analysis (PCA) on the significantly up-regulated genes and using the first component of the PCA as the score (80), according to the equation
Here, X represents the gene expression matrix size n x m, where n are the signature genes and m are the cells. U and V are m × m and n × n orthogonal matrices, respectively, and Σ is a rectangular diagonal matrix. We then used the first column of U as a signature score. We then ranked the cell populations based on the difference in the mean score between the WT and the STINGN153S mutant. Gene set enrichment was performed with g:profiler (package gprofiler-offical, version 1.0.0) (81) on all GO:BP (releases/2021-05-01) terms that were enriched for more than 400 genes. Filtering on gene set size was performed to exclude terms that were too general. All genes expressed in the dataset were used as the background.
Statistical analyses
Statistical analyses were performed using GraphPad Prism. The statistical tests are described in the respective figure legends. In short, unpaired two-tailed Student’s t test (comparison of two groups), ordinary one-way analysis of variance (ANOVA) combined with Dunnett’s multiple comparison test (comparison of more than two groups), and log-rank (Mantel-Cox) test were used. The data are presented as the mean ± SD if not stated otherwise. P < 0.05 was considered to indicate statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Acknowledgments
We would like to thank V. Höfl, K. Burmeister, L. Bachmann, P. Konstantinidis, and J. Höbart for technical assistance; R. Öllinger and R. Rad for assistance in RNA-seq; T. Walzthöni for assistance in CITE-seq; and the data analysis team from AG Theis for discussions and feedback. The cell lines for the production of R-spondin 1 CM and Noggin CM were gifts from C. Kuo and H. Clevers, respectively. Medical Art by Servier was used to create figures in this article, as licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com).
Funding: J.R. was supported by research grants from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) (Project-ID 210592381–SFB 1054, Project-ID 360372040–SFB 1335, Project-ID 395357507–SFB 1371, Project-ID 369799452–TRR 237, Project-ID 452881907–TRR 338, Project-ID 435874434–RTG 2668, RU 695/9-1, and RU 695/12-1), the Leukemia & Lymphoma Society, and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 834154). L.V. received funding from the international doctoral program “i-Target: Immunotargeting in Cancer,” funded by the Elite Network of Bavaria. Work in the laboratories of H.F.F. and F.R.G. was supported by institutional funds from the Georg-Speyer-Haus and the LOEWE Center Frankfurt Cancer Institute (FCI) funded by the Hessen State Ministry for Higher Education, Research and the Arts [III L 5-519/03/03.001-(0015)] as well as the Deutsche Forschungsgemeinschaft (FOR2438: FA 1334/2-2). The Institute for Tumor Biology and Experimental Therapy, Georg-Speyer-Haus, is funded jointly by the German Federal Ministry of Health and the Ministry of Higher Education, Research and the Arts of the State of Hessen (HMWK).
Author contributions: Conceptualization: L.V. and J.R. Investigation: L.V., S.E.I., Z.K., D.C.S., P.L., M.H.M., M.D.L., M.S., and C.W. Funding acquisition: H.L., F.R.G., H.F.F., F.J.T., and J.R. Writing: L.V. and J.R. All authors discussed the results and reviewed the manuscript.
Competing interests: M.D.L. contracted for the Chan Zuckerberg Initiative and received speaker fees from Pfizer and Janssen Pharmaceuticals. F.J.T. consults for Immunai Inc., Singularity Bio B.V., CytoReason Ltd., and Omniscope Ltd. and has ownership interest in Dermagnostix GmbH and Cellarity. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Patient-derived organoids can be shared upon reasonable request to H.F.F. (h.farin@georg-speyer-haus.de), following approval by the local ethical review board of the University Cancer Center Frankfurt. The MC38 STINGN153S RNA-seq and CITE-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) (82) and are accessible through GEO Series accession number GSE216898. Proprietary cell lines are available from the corresponding author upon reasonable request with a material transfer agreement.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legends for tables S1 to S3
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.C. Robert, A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 11, 3801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D. S. Chen, I. Mellman, Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 3.J. Galon, D. Bruni, Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 18, 197–218 (2019). [DOI] [PubMed] [Google Scholar]
- 4.D. T. Le, J. N. Durham, K. N. Smith, H. Wang, B. R. Bartlett, L. K. Aulakh, S. Lu, H. Kemberling, C. Wilt, B. S. Luber, F. Wong, N. S. Azad, A. A. Rucki, D. Laheru, R. Donehower, A. Zaheer, G. A. Fisher, T. S. Crocenzi, J. J. Lee, T. F. Greten, A. G. Duffy, K. K. Ciombor, A. D. Eyring, B. H. Lam, A. Joe, S. P. Kang, M. Holdhoff, L. Danilova, L. Cope, C. Meyer, S. Zhou, R. M. Goldberg, D. K. Armstrong, K. M. Bever, A. N. Fader, J. Taube, F. Housseau, D. Spetzler, N. Xiao, D. M. Pardoll, N. Papadopoulos, K. W. Kinzler, J. R. Eshleman, B. Vogelstein, R. A. Anders, L. A. Diaz Jr., Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.G.-Y. Lyu, Y.-H. Yeh, Y.-C. Yeh, Y.-C. Wang, Mutation load estimation model as a predictor of the response to cancer immunotherapy. NPJ Genom. Med. 3, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.X. Meng, Z. Huang, F. Teng, L. Xing, J. Yu, Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat. Rev. 41, 868–876 (2015). [DOI] [PubMed] [Google Scholar]
- 7.P. Mur, S. García-Mulero, J. del Valle, L. Magraner-Pardo, A. Vidal, M. Pineda, G. Cinnirella, E. Martín-Ramos, T. Pons, A. López-Doriga, S. Belhadj, L. Feliubadaló, P. M. Munoz-Torres, M. Navarro, E. Grau, E. Darder, G. Llort, J. Sanz, T. Ramón y Cajal, J. Balmana, J. Brunet, V. Moreno, J. M. Piulats, X. Matías-Guiu, R. Sanz-Pamplona, R. Aligué, G. Capellá, C. Lázaro, L. Valle, Role of POLE and POLD1 in familial cancer. Genet. Med. 22, 2089–2100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.N. Pećina-Šlaus, A. Kafka, I. Salamon, A. Bukovac, Mismatch repair pathway, genome stability and cancer. Front. Mol. Biosci. 7, 122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D. Hanahan, R. A. Weinberg, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 10.C. R. Boland, A. Goel, Microsatellite instability in colorectal cancer. Gastroenterology 138, 2073–2087.e3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.G. Germano, S. Lamba, G. Rospo, L. Barault, A. Magrì, F. Maione, M. Russo, G. Crisafulli, A. Bartolini, G. Lerda, G. Siravegna, B. Mussolin, R. Frapolli, M. Montone, F. Morano, F. de Braud, N. Amirouchene-Angelozzi, S. Marsoni, M. D’Incalci, A. Orlandi, E. Giraudo, A. Sartore-Bianchi, S. Siena, F. Pietrantonio, F. Di Nicolantonio, A. Bardelli, Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120 (2017). [DOI] [PubMed] [Google Scholar]
- 12.G. Poulogiannis, I. M. Frayling, M. J. Arends, DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 56, 167–179 (2010). [DOI] [PubMed] [Google Scholar]
- 13.GBD 2017 Colorectal Cancer Collaborators , The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 4, 913–933 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. L. Siegel, K. D. Miller, A. Goding Sauer, S. A. Fedewa, L. F. Butterly, J. C. Anderson, A. Cercek, R. A. Smith, A. Jemal, Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164 (2020). [DOI] [PubMed] [Google Scholar]
- 15.R. Bonneville, M. A. Krook, E. A. Kautto, J. Miya, M. R. Wing, H.-Z. Chen, J. W. Reeser, L. Yu, S. Roychowdhury, Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Umar, C. R. Boland, J. P. Terdiman, S. Syngal, A. de la Chapelle, J. Rüschoff, R. Fishel, N. M. Lindor, L. J. Burgart, R. Hamelin, S. R. Hamilton, R. A. Hiatt, J. Jass, A. Lindblom, H. T. Lynch, P. Peltomaki, S. D. Ramsey, M. A. Rodriguez-Bigas, H. F. A. Vasen, E. T. Hawk, J. C. Barrett, A. N. Freedman, S. Srivastava, Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 96, 261–268 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. T. Le, J. N. Uram, H. Wang, B. R. Bartlett, H. Kemberling, A. D. Eyring, A. D. Skora, B. S. Luber, N. S. Azad, D. Laheru, B. Biedrzycki, R. C. Donehower, A. Zaheer, G. A. Fisher, T. S. Crocenzi, J. J. Lee, S. M. Duffy, R. M. Goldberg, A. de la Chapelle, M. Koshiji, F. Bhaijee, T. Huebner, R. H. Hruban, L. D. Wood, N. Cuka, D. M. Pardoll, N. Papadopoulos, K. W. Kinzler, S. Zhou, T. C. Cornish, J. M. Taube, R. A. Anders, J. R. Eshleman, B. Vogelstein, L. A. Diaz Jr., PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R. Mandal, R. M. Samstein, K.-W. Lee, J. J. Havel, H. Wang, C. Krishna, E. Y. Sabio, V. Makarov, F. Kuo, P. Blecua, A. T. Ramaswamy, J. N. Durham, B. Bartlett, X. Ma, R. Srivastava, S. Middha, A. Zehir, J. F. Hechtman, L. G. Morris, N. Weinhold, N. Riaz, D. T. Le, L. A. Diaz Jr., T. A. Chan, Genetic diversity of tumors with mismatch repair deficiency influences anti–PD-1 immunotherapy response. Science 364, 485–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.C. Lu, J. Guan, S. Lu, Q. Jin, B. Rousseau, T. Lu, D. Stephens, H. Zhang, J. Zhu, M. Yang, Z. Ren, Y. Liang, Z. Liu, C. Han, L. Liu, X. Cao, A. Zhang, J. Qiao, K. Batten, M. Chen, D. H. Castrillon, T. Wang, B. Li, L. A. Diaz Jr., G.-M. Li, Y.-X. Fu, DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 39, 96–108.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.K. J. Mackenzie, P. Carroll, C.-A. Martin, O. Murina, A. Fluteau, D. J. Simpson, N. Olova, H. Sutcliffe, J. K. Rainger, A. Leitch, R. T. Osborn, A. P. Wheeler, M. Nowotny, N. Gilbert, T. Chandra, M. A. M. Reijns, A. P. Jackson, cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.F. Talens, M. A. T. M. Van Vugt, Inflammatory signaling in genomically instable cancers. Cell Cycle 18, 1830–1848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.L. Sun, J. Wu, F. Du, X. Chen, Z. J. Chen, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.K.-P. Hopfner, V. Hornung, Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Y.-C. Perng, D. J. Lenschow, ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 16, 423–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.T. J. Zumwalt, M. Arnold, A. Goel, C. R. Boland, Active secretion of CXCL10 and CCL5 from colorectal cancer microenvironments associates with Granzyme B+ CD8+ T-cell infiltration. Oncotarget 6, 2981–2991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. P. Cullen, M. Brunet, S. J. Martin, Granzymes in cancer and immunity. Cell Death Differ. 17, 616–623 (2010). [DOI] [PubMed] [Google Scholar]
- 27.W. Du, T. L. Frankel, M. Green, W. Zou, IFNγ signaling integrity in colorectal cancer immunity and immunotherapy. Cell. Mol. Immunol. 19, 23–32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Y. Liu, A. A. Jesus, B. Marrero, D. Yang, S. E. Ramsey, G. A. M. Sanchez, K. Tenbrock, H. Wittkowski, O. Y. Jones, H. S. Kuehn, C.-C. R. Lee, M. A. DiMattia, E. W. Cowen, B. Gonzalez, I. Palmer, J. J. DiGiovanna, A. Biancotto, H. Kim, W. L. Tsai, A. M. Trier, Y. Huang, D. L. Stone, S. Hill, H. J. Kim, C. St. Hilaire, S. Gurprasad, N. Plass, D. Chapelle, I. Horkayne-Szakaly, D. Foell, A. Barysenka, F. Candotti, S. M. Holland, J. D. Hughes, H. Mehmet, A. C. Issekutz, M. Raffeld, J. McElwee, J. R. Fontana, C. P. Minniti, S. Moir, D. L. Kastner, M. Gadina, A. C. Steven, P. T. Wingfield, S. R. Brooks, S. D. Rosenzweig, T. A. Fleisher, Z. Deng, M. Boehm, A. S. Paller, R. Goldbach-Mansky, Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371, 507–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.H. Luksch, W. A. Stinson, D. J. Platt, W. Qian, G. Kalugotla, C. A. Miner, B. G. Bennion, A. Gerbaulet, A. Rösen-Wolff, J. J. Miner, STING-associated lung disease in mice relies on T cells but not type I interferon. J. Allergy Clin. Immunol. 144, 254–266.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.W. M. Schneider, M. D. Chevillotte, C. M. Rice, Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S. Liu, X. Cai, J. Wu, Q. Cong, X. Chen, T. Li, F. Du, J. Ren, Y.-T. Wu, N. V. Grishin, Z. J. Chen, Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 32.W. A. Hudson, Q. Li, C. Le, J. H. Kersey, Xenotransplantation of human lymphoid malignancies is optimized in mice with multiple immunologic defects. Leukemia 12, 2029–2033 (1998). [DOI] [PubMed] [Google Scholar]
- 33.H. Bronger, J. Singer, C. Windmüller, U. Reuning, D. Zech, C. Delbridge, J. Dorn, M. Kiechle, B. Schmalfeldt, M. Schmitt, S. Avril, CXCL9 and CXCL10 predict survival and are regulated by cyclooxygenase inhibition in advanced serous ovarian cancer. Br. J. Cancer 115, 553–563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Y. Cao, N. Jiao, T. Sun, Y. Ma, X. Zhang, H. Chen, J. Hong, Y. Zhang, CXCL11 correlates with antitumor immunity and an improved prognosis in colon cancer. Front. Cell Dev. Biol. 9, 646252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. B. Fuertes, A. K. Kacha, J. Kline, S.-R. Woo, D. M. Kranz, K. M. Murphy, T. F. Gajewski, Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells. J. Exp. Med. 208, 2005–2016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R. Noubade, S. Majri-Morrison, K. V. Tarbell, Beyond cDC1: Emerging roles of DC crosstalk in cancer immunity. Front. Immunol. 10, 1014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Stoeckius, C. Hafemeister, W. Stephenson, B. Houck-Loomis, P. K. Chattopadhyay, H. Swerdlow, R. Satija, P. Smibert, Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Gayoso, Z. Steier, R. Lopez, J. Regier, K. L. Nazor, A. Streets, N. Yosef, Joint probabilistic modeling of single-cell multi-omic data with total VI. Nat. Methods 18, 272–282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Guan, C. Lu, Q. Jin, H. Lu, X. Chen, L. Tian, Y. Zhang, J. Ortega, J. Zhang, S. Siteni, M. Chen, L. Gu, J. W. Shay, A. J. Davis, Z. J. Chen, Y.-X. Fu, G.-M. Li, MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS-STING pathway. Cancer Cell 39, 109–121.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. S. Garris, S. P. Arlauckas, R. H. Kohler, M. P. Trefny, S. Garren, C. Piot, C. Engblom, C. Pfirschke, M. Siwicki, J. Gungabeesoon, G. J. Freeman, S. E. Warren, S. Ong, E. Browning, C. G. Twitty, R. H. Pierce, M. H. Le, A. P. Algazi, A. I. Daud, S. I. Pai, A. Zippelius, R. Weissleder, M. J. Pittet, Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity 49, 1148–1161.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.B. A. Flood, E. F. Higgs, S. Li, J. J. Luke, T. F. Gajewski, STING pathway agonism as a cancer therapeutic. Immunol. Rev. 290, 24–38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L. Motedayen Aval, J. E. Pease, R. Sharma, D. J. Pinato, Challenges and opportunities in the clinical development of STING agonists for cancer immunotherapy. J. Clin. Med. 9, 3323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. Su, Y. Zhang, K. Valerie, X.-Y. Wang, S. Lin, G. Zhu, STING activation in cancer immunotherapy. Theranostics 9, 7759–7771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. Cerboni, N. Jeremiah, M. Gentili, U. Gehrmann, C. Conrad, M.-C. Stolzenberg, C. Picard, B. Neven, A. Fischer, S. Amigorena, F. Rieux-Laucat, N. Manel, Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214, 1769–1785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. F. Gulen, U. Koch, S. M. Haag, F. Schuler, L. Apetoh, A. Villunger, F. Radtke, A. Ablasser, Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8, 427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.B. Larkin, V. Ilyukha, M. Sorokin, A. Buzdin, E. Vannier, A. Poltorak, Cutting edge: Activation of STING in T cells induces type I IFN responses and cell death. J. Immunol. 199, 397–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. Wu, N. Dobbs, K. Yang, N. Yan, Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity 53, 115–126.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.H. Lemos, E. Mohamed, L. Huang, R. Ou, G. Pacholczyk, A. S. Arbab, D. Munn, A. L. Mellor, STING promotes the growth of tumors characterized by low antigenicity via IDO activation. Cancer Res. 76, 2076–2081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. Ahn, T. Xia, H. Konno, K. Konno, P. Ruiz, G. N. Barber, Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.H. Liang, L. Deng, Y. Hou, X. Meng, X. Huang, E. Rao, W. Zheng, H. Mauceri, M. Mack, M. Xu, Y.-X. Fu, R. R. Weichselbaum, Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat. Commun. 8, 1736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.H. Konno, S. Yamauchi, A. Berglund, R. M. Putney, J. J. Mulé, G. N. Barber, Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 37, 2037–2051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. Song, P. Peng, Z. Tang, J. Zhao, W. Wu, H. Li, M. Shao, L. Li, C. Yang, F. Duan, M. Zhang, J. Zhang, H. Wu, C. Li, X. Wang, H. Wang, Y. Ruan, J. Gu, Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci. Rep. 7, 39858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.T. Xia, H. Konno, G. N. Barber, Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 76, 6747–6759 (2016). [DOI] [PubMed] [Google Scholar]
- 54.T. Xia, H. Konno, J. Ahn, G. N. Barber, Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14, 282–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Le Naour, L. Zitvogel, L. Galluzzi, E. Vacchelli, G. Kroemer, Trial watch: STING agonists in cancer therapy. Oncoimmunology 9, 1777624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. E. Lawler, M.-C. Speranza, C.-F. Cho, E. A. Chiocca, Oncolytic viruses in cancer treatment: A review. JAMA Oncol. 3, 841–849 (2017). [DOI] [PubMed] [Google Scholar]
- 57.C. Hotz, T. R. Wagenaar, F. Gieseke, D. S. Bangari, M. Callahan, H. Cao, J. Diekmann, M. Diken, C. Grunwitz, A. Hebert, K. Hsu, M. Bernardo, K. Karikó, S. Kreiter, A. N. Kuhn, M. Levit, N. Malkova, S. Masciari, J. Pollard, H. Qu, S. Ryan, A. Selmi, J. Schlereth, K. Singh, F. Sun, B. Tillmann, T. Tolstykh, W. Weber, L. Wicke, S. Witzel, Q. Yu, Y.-A. Zhang, G. Zheng, J. Lager, G. J. Nabel, U. Sahin, D. Wiederschain, Local delivery of mRNA-encoded cytokines promotes antitumor immunity and tumor eradication across multiple preclinical tumor models. Sci. Transl. Med. 13, eabc7804 (2021). [DOI] [PubMed] [Google Scholar]
- 58.S.-W. Tse, K. McKinney, W. Walker, M. Nguyen, J. Iacovelli, C. Small, K. Hopson, T. Zaks, E. Huang, mRNA-encoded, constitutively active STINGV155M is a potent genetic adjuvant of antigen-specific CD8+ T cell response. Mol. Ther. 29, 2227–2238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.L. Miao, L. Li, Y. Huang, D. Delcassian, J. Chahal, J. Han, Y. Shi, K. Sadtler, W. Gao, J. Lin, J. C. Doloff, R. Langer, D. G. Anderson, Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019). [DOI] [PubMed] [Google Scholar]
- 60.J. G. Rurik, I. Tombácz, A. Yadegari, P. O. Méndez Fernández, S. V. Shewale, L. Li, T. Kimura, O. Y. Soliman, T. E. Papp, Y. K. Tam, B. L. Mui, S. M. Albelda, E. Puré, C. H. June, H. Aghajanian, D. Weissman, H. Parhiz, J. A. Epstein, CAR T cells produced in vivo to treat cardiac injury. Science 375, 91–96 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.G. A. Van der Auwera, M. O. Carneiro, C. Hartl, R. Poplin, G. Del Angel, A. Levy-Moonshine, T. Jordan, K. Shakir, D. Roazen, J. Thibault, E. Banks, K. V. Garimella, D. Altshuler, S. Gabriel, M. A. DePristo, From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.K. Cibulskis, M. S. Lawrence, S. L. Carter, A. Sivachenko, D. Jaffe, C. Sougnez, S. Gabriel, M. Meyerson, E. S. Lander, G. Getz, Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.E. Reble, C. A. Castellani, M. G. Melka, R. O’Reilly, S. M. Singh, VarScan2 analysis of de novo variants in monozygotic twins discordant for schizophrenia. Psychiatr. Genet. 27, 62–70 (2017). [DOI] [PubMed] [Google Scholar]
- 64.W. McLaren, L. Gil, S. E. Hunt, H. S. Riat, G. R. S. Ritchie, A. Thormann, P. Flicek, F. Cunningham, The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.G. Korotkevich, V. Sukhov, N. Budin, B. Shpak, M. N. Artyomov, A. Sergushichev, Fast gene set enrichment analysis. bioRxiv 10.1101/060012 [Preprint]. 1 February 2021. 10.1101/060012. [DOI]
- 66.A. Schambach, J. Bohne, S. Chandra, E. Will, G. P. Margison, D. A. Williams, C. Baum, Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine–DNA methyltransferase in hematopoietic cells. Mol. Ther. 13, 391–400 (2006). [DOI] [PubMed] [Google Scholar]
- 67.T. E. Schnalzger, M. H. de Groot, C. Zhang, M. H. Mosa, B. E. Michels, J. Röder, T. Darvishi, W. S. Wels, H. F. Farin, 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 38, e100928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.S. Parekh, C. Ziegenhain, B. Vieth, W. Enard, I. Hellmann, The impact of amplification on differential expression analyses by RNA-seq. Sci. Rep. 6, 25533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev, S. A. McCarroll, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Z. Fang, X. Liu, G. Peltz, GSEApy: A comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 39, btac757 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.F. A. Wolf, P. Angerer, F. J. Theis, SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.M. D. Luecken, F. J. Theis, Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 15, e8746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.A. T. L. Lun, D. J. McCarthy, J. C. Marioni, A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 31, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.L. McInnes, J. Healy, J. Melville, UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426 [stat.ML] (18 September 2020).
- 77.V. A. Traag, L. Waltman, N. J. van Eck, From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.theislab/diffxpy (2022); https://github.com/theislab/diffxpy.
- 79.Y. Benjamini, Y. Hochberg, Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995). [Google Scholar]
- 80.P. Langfelder, S. Horvath, Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 1, 54 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.U. Raudvere, L. Kolberg, I. Kuzmin, T. Arak, P. Adler, H. Peterson, J. Vilo, g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R. Edgar, M. Domrachev, A. E. Lash, Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for tables S1 to S3
Tables S1 to S3