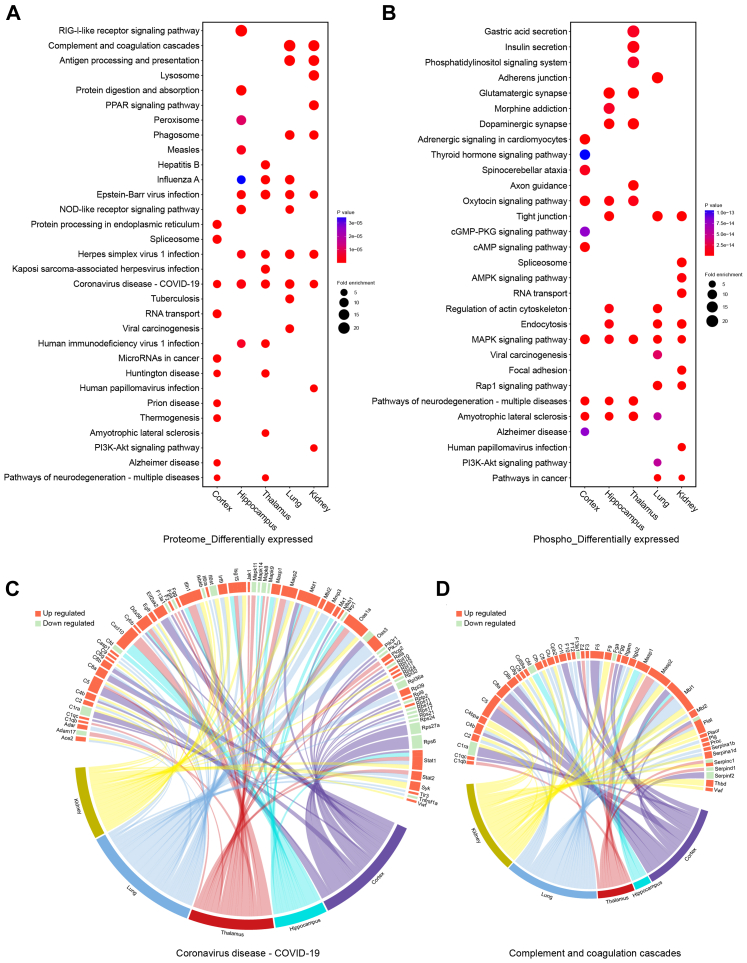
Fig. 2.
Functional analyses for differentially expressed proteins in the proteome and proteins containing differentially expressed phosphosites in the phosphoproteome. KEGG pathways enriched for the cortex, hippocampus, thalamus, lung, and kidney are integrated into bubble plots. A. Functional enrichment analyses for differentially expressed proteins in the five organs. Fold change (FC) and P value of Student's t test were separately calculated in each organ. That is, FCprotein = (mean value of four infected samples)/(mean value of corresponding four control samples) and each organ had differentially expressed proteins independent of other organs. Differentially expressed proteins contained both up- and downregulated proteins. Upregulated proteins were defined as FCprotein >1.5 and P < 0.05 (Student's t test, n = 4). Downregulated proteins were defined as FCprotein <1/1.5 and P < 0.05 (Student's t test, n = 4). The top 10 KEGG pathways enriched in each organ were integrated and presented in the bubble plots. B. Functional enrichment analyses for proteins containing differentially expressed phosphosites in the five organs. FC and P value of Student's t test were separately calculated in each organ. FCphosphosite = (mean value of four infected samples)/(mean value of corresponding four control samples) and each organ had differentially expressed phosphosites independent of other organs. Differentially expressed phosphosites contained both up- and downregulated phosphosites. Upregulated phosphosites were defined as FCphosphosite >1.5 and P < 0.05 (Student's t test, n = 4). Downregulated phosphosites were defined as FCphosphosite <1/1.5 and P < 0.05 (Student's t test, n = 4). Proteins containing differentially expressed, upregulated, or downregulated phosphosites were used for functional enrichment analysis. The top 10 KEGG pathways enriched in each organ are integrated and presented in the bubble plots. C. Circos plot showing the correspondence between organs and differentially expressed proteins in a KEGG pathway. Differentially expressed proteins belonging to the COVID-19 pathway in each organ were presented in the plot. Red segments show upregulated proteins and green segments show downregulated proteins. The length of each segment corresponds to the sum of |log2 FC| of each protein. D. Circos plot showing the correspondence between organs and differentially expressed proteins in a KEGG pathway. Differentially expressed proteins belonging to complement and coagulation cascades in each organ are presented in the plot. Red segments show upregulated proteins and green segments show downregulated proteins. The length of each segment corresponds to the sum of |log2 FC| of each protein.