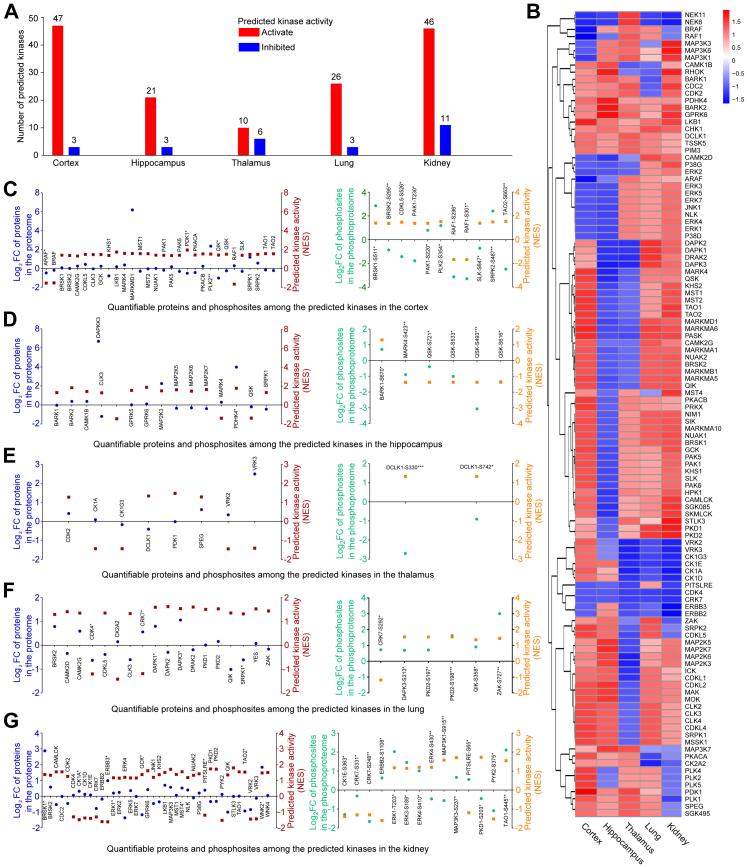
Fig. 4.
Kinase prediction and kinase activity prediction based on the phosphoproteome. A. Number of predicted kinases in each organ after SARS-CoV-2 infection. Based on quantification of phosphosites, kinase prediction was performed using the iGPS algorithm and kinase activity prediction was performed using the GSEA algorithm. NES values derived from the GSEA algorithm reflect predicted kinase activity. The threshold of predicted activated kinases is set at NES >0. The threshold of predicated inhibited kinases is set at NES <0. B. Heat map of predicted kinases that may have had significantly altered activity after SARS-CoV-2 infection. Kinase activity is represented by NES values. C–G. Comparison of the predicted activity of kinases and actual measured FC of proteins and phosphosites in the cortex (C), hippocampus (D), thalamus (E), lung (F), and kidney (G) after SARS-CoV-2 infection (n = 4, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Student's t test)). The predicted kinases were matched to quantifiable proteins or phosphosites in each organ. The left panel shows quantification of alterations of proteins in the proteome and the corresponding kinase activity. The right panel shows quantification of alterations of phosphosites in the phosphoproteome and the corresponding kinase activity. Red and orange squares represent log2 FC of protein or phosphosites, respectively. Blue and green points represent the NES of the kinases.