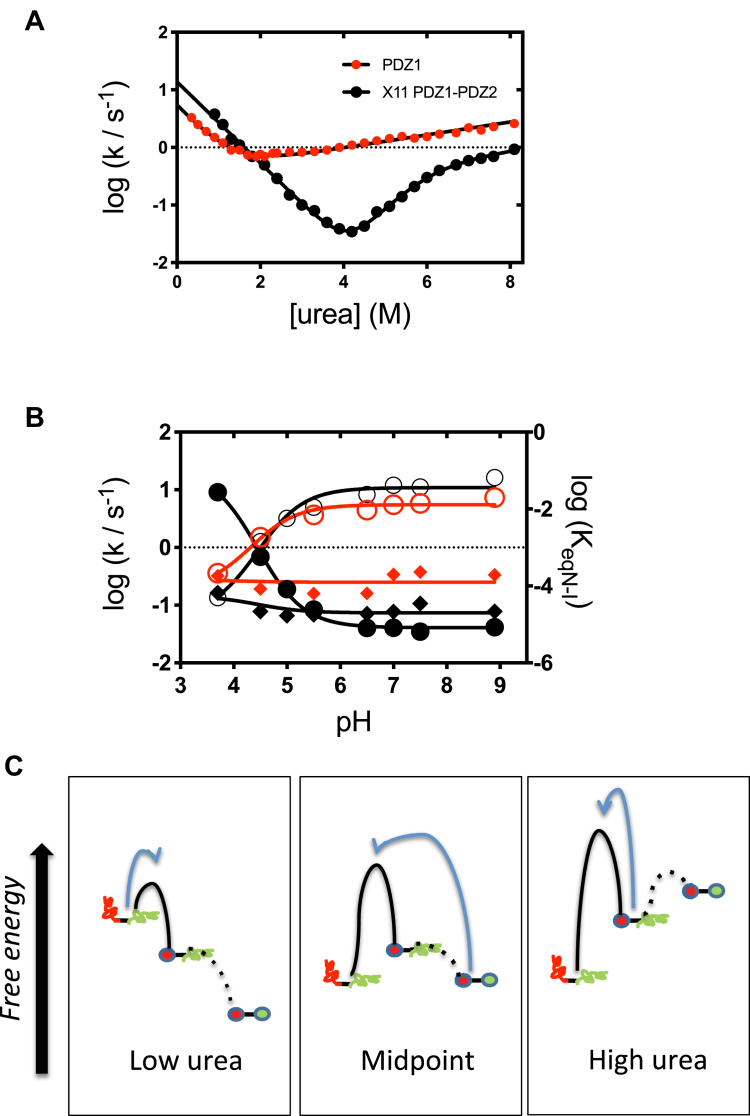
Figure 3.
Folding kinetics of X11 PDZ1 and PDZ2.A, the chevron plots of X11 PDZ1-PDZ2, black circles, and PDZ1, red circles. It is evident that, while X11 PDZ1-PDZ2 presents a pronounced curvature in the unfolding branch, PDZ1 conforms to a V-shape chevron plot. Accordingly, the two different constructs were fitted to a three-state and a two-state model, respectively. Lines are the best fit obtained for the two proteins. B, dependence of the folding and unfolding rate constants of X11 PDZ1-PDZ2 and PDZ1 on pH. The folding rate constants kF are represented in open circles; unfolding rate constants kU are represented in diamonds, whereas the stability of the folding intermediate as probed by the KeqN–I, which could be identified for X11 PDZ1-PDZ2 only, is represented in filled circles. By following a three-state analysis of the observed chevron plots, the KeqN–I, value was estimated in the absence of denaturant, and the calculated values are reported in the right-y axis. Parameters referring to X11 PDZ1.PDZ2 are reported in black, whereas those referring to PDZ1 are reported in red. As discussed in the test, it is evident that X11 PDZ1-PDZ2 and PDZ1 display a very similar pH dependence of the folding and unfolding rate constants, indicating that at very high and low denaturant concentrations, (un)folding is limited by PDZ1. This finding is further supported by the conserved m values for these two parameters at different experimental conditions (see text and Fig. S1). C, a plausible energy diagram describing the reaction mechanism of (un)folding of X11 PDZ1-PDZ2. PDZ1 is depicted in red, whereas PDZ2 is depicted in green. The diagram schematically explains how both folding at low denaturant concentrations and unfolding at very high denaturant concentrations are limited by PDZ1. By following this view, the folding intermediate should involve the presence of folded PDZ1 with PDZ2 by-and-large denatured.