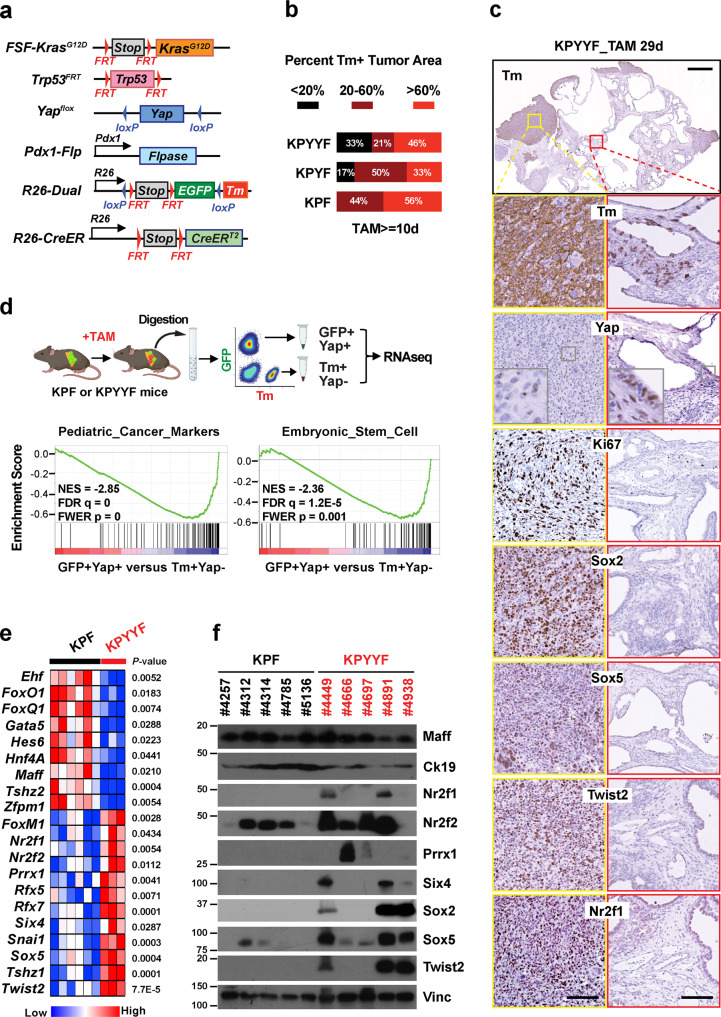
Fig. 1. A subset of Kras:p53 mutant PDAC tumors develop spontaneous relapsed lesions with embryonic progenitor features following Yap ablation.
a Genetic strategy to first activate KrasG12D and delete Trp53 in the pancreas via Pdx1-Flp, and subsequently delete Yap via TAM-induced Cre-loxP recombination systems. Note that FSF-KrasG12D/Trp53FRT and Yapflox/flox are under the separate controls of Flp and CreER, respectively. The R26-Dual reporter marks Flp-expressing KrasG12D cells with EGFP. Upon TAM-mediated CreER activation, the EGFP locus is removed, while tdTomato (Tm) is switched on. b The percentage of recombination evaluated by Tm+ area over Tm+ and GFP+ area based on IHC staining of KPF (n = 9), KPYF (n = 6) and KPYYF (n = 33) PDAC tumors following TAM treatment for at least 10 days. c Representative images of IHC analysis of indicated proteins in a KPYYF PDAC tumor after 29 days of TAM treatment. The top panel shows the overall Tm staining of an entire tumor with two small boxes marking a representative relapsing region (yellow) and a representative regressing region (red), respectively. The bottom panels include the higher-magnification IHC images of the indicated proteins at the marked regions. Scale bars indicate 1 mm (top) or 50 μm (all other panels). Experiments were performed on 13 different KPYY mice. d Experimental design (top) and the top two most enriched Gene Sets in Tm+/Yap− relative to GFP+/Yap+ cells according to GSEA (bottom) of RNA-seq of PDAC cells isolated from TAM-treated KPF and KPYYF mice. Unpaired, One-sided, Z-test. Created with BioRender.com. e Heatmap representing the expression of indicated genes in KPF (n = 6) and KPYYF (n = 3) tumors. Unpaired, two-tailed, Student’s t-test. f Western blot (WB) analysis of indicated proteins in KPF and KPYYF tumors. Vinc was used as the loading control. The mouse IDs belonging to the KPF or KPYYF cohorts are indicated. Shown is representative of three independent experiments. Source data are provided within a Source Data file.