SUMMARY
The transfer of endocytosed cargoes to lysosomes (LYSs) requires HOPS, a multiprotein complex that tethers late endosomes (LEs) to LYSs before fusion. Many proteins interact with HOPS on LEs/LYSs. However, it is not clear whether these HOPS interactors localize to LEs or LYSs or how they participate in tethering. Here, we biochemically characterized endosomes purified from untreated or experimentally manipulated cells to put HOPS and interacting proteins in order and to establish their functional interdependence. Our results assign Rab2a and Rab7 to LEs and Arl8 and BORC to LYSs and show that HOPS drives LE-LYS fusion by bridging late endosomal Rab2a with lysosomal BORC-anchored Arl8. We further show that Rab7 is absent from sites of HOPS-dependent tethering but promotes fusion by moving LEs toward LYSs via dynein. Thus, our study identifies the topology of the machinery for LE-LYS tethering and elucidates the role of different small GTPases in the process.
Graphical Abstract
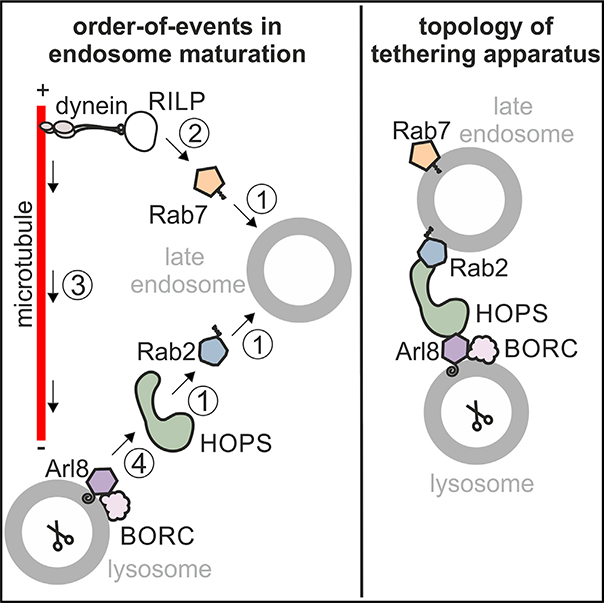
In brief
Schleinitz et al. identify the order of action of small GTPases in endosome maturation and characterize the topology of the apparatus that drives late endosome-lysosome fusion: the HOPS tethering complex guides fusion by bridging Rab2 on late endosomes with Arl8 on lysosomes.
INTRODUCTION
Endocytosed cargoes pass through early (EEs) and late endosomes (LEs) before being degraded by lysosomal acid hydrolases in endolysosomes (ELYSs).1 EEs localize to the cell periphery and segregate proteins to be recycled from macromolecules to be degraded.2 EEs mature into LEs3 and/or generate endosomal carrier vesicles that deliver endocytosed cargoes to pre-existing LEs by membrane fusion.4 LEs move along microtubules toward the perinuclear microtubule-organizing center (MTOC) and sequester transmembrane proteins marked for degradation into intraluminal vesicles (ILVs).5
LEs acidify and eventually fuse with lysosomes (LYSs), resulting in the formation of ELYSs, i.e., LE-LYS hybrids in which soluble and ILV cargoes are degraded by mature lysosomal hydrolases.1,5 ELYSs are transient compartments from which LYSs can be reformed.1
Cargo delivery to LYSs requires that LEs and LYSs are transported toward each other and that they contain a cognate set of protein and lipid factors for fusion. The microtubule-dependent transport and recruitment of fusion factors to LEs/LYSs is coordinated by the small GTPases Ras-like protein from rat brain 7 (Rab7) and Arf-like GTPase 8 (Arl8), which exists as two paralogs named Arl8a and Arl8b.6 Rab7 and Arl8 cycle between a GTP-bound, membrane-associated, active and a GDP-bound, largely cytosolic, inactive conformation.7,8 When activated, these GTPases recruit effector proteins to membranes.9
Rab7 itself is recruited to EEs and activated by the monensin sensitivity 1a/calcium caffeine zinc sensitivity 1 (Mon1a/Ccz1) complex that has been recruited to EEs and activated by Rab5(GTP).10 Mon1a/Ccz1 displaces the Rab5-guanine nucleotide exchange factor (GEF) Rabex5 from membranes, acts as a GEF for Rab7,10–12 and causes a Rab5-to-Rab7 conversion between EEs and LEs.3,13
Arl8 is recruited to LEs/LYSs by BLOC one-related complex (BORC), a multi-protein complex composed of BLOS1, BLOS2, snapin, KXD1, myrlysin, lyspersin, diaskedin, and MEF2BNB that may act as an Arl8-GEF, an Arl8-GEF adaptor, and/or a membrane receptor of Arl8.14,15
Rab7 promotes endosome transport toward microtubule minus ends at the perinuclear MTOC by recruiting the dynein-dynactin motor protein complex through its effectors Rab-interacting lysosomal protein (RILP) and oxysterol-binding protein-related protein 1L (ORP1L).16 Arl8, by contrast, promotes transport of late endocytic compartments toward microtubule plus ends in the cell periphery by recruiting kinesin-1 through its effector SifA- and kinesin-interacting protein (SKIP)17,18 and kinesin-3 directly.19 These antagonistic activities may help to transport Rab7- and Arl8-containing compartments toward each other, facilitating their encounter and fusion.20
Besides their roles in endosome transport, Arl8 directly7 and Rab7 indirectly21,22 anchor the homotypic fusion and vacuolar protein sorting (HOPS) complex to membranes. HOPS is a multi-subunit tethering complex that, in the yeast endocytic system, guides tethering, trans-SNARE complex formation, and fusion between LYSs.23 Mammalian HOPS binds to the Rab7 effector RILP21,22; to Arl87,24; to the dual Rab7/Arl8-effectors pleckstrin homology domain-containing family M member 1 (Plekhm16,25) and SKIP7,26; to Rab2a27 and Rab2b28; to late endocytic SNARE proteins29–31; and possibly to BORC20 (Figure S1A). Thus, there are many possibilities for how HOPS could be anchored to adjacent LEs and/or LYSs before fusion. For example, HOPS may (1) link two copies of Rab7 via RILP on both ends of HOPS21 (Figure S1B, 1); (2) Rab7 and Arl8 on opposing membranes via Rab7-RILP on one membrane and Arl8 on the other6 (Figure S1B, 2); or (3) Rab2 on one compartment and Arl8 on the other32 (Figure S1B, 3). If Plekhm1 and SKIP contribute to HOPS-dependent tethering is unclear. Plekhm1 binds to Arl8 and Rab7 on adjacent membranes6 (Figure S1B, 4), whereas SKIP recognizes Arl8 and Rab7 present on the same membrane.26 Hence, Plekhm1 is more likely to contribute to late endocytic tethering with or without HOPS. An explanation for how HOPS is anchored to LEs/LYSs is further complicated by the fact that HOPS may bind to phosphatidylinositol phosphates (PIPs).33,34
It has been speculated that Rab7 resides on endocytic compartments of an earlier maturation stage than Arl8.26,35 Both Arl8 and Rab7 colocalize with the LE/LYS protein LYS-associated membrane protein 1 (LAMP1)7 and to some extent with each other. Arl8 recruits the Rab7- GTPase-activating protein (GAP) TBC1 domain family member 15 (TBC1D15) via SKIP and HOPS, resulting in the removal of Rab7 from Arl8-containing compartments.26 Thus, apparently, endosome maturation encompasses a Rab7-to-Arl8 switch downstream of the Rab5-to-Rab7 switch between EEs and LEs.26
Although much work has been dedicated to identifying the roles of Rab7 and Arl8 and their various effectors and the multiple interactions between them, it is unclear in which order these factors act.6 Moreover, it is not known which proteins anchor HOPS on LEs and LYSs.
We now report that HOPS links Rab2a on LEs with BORC-anchored Arl8 on LYSs and that Rab7 does not participate in HOPS-dependent tethering yet stimulates fusion by moving LEs toward LYSs via dynein.
RESULTS
We analyzed endosomes from HeLa cells isolated after endocytosis of 10 nm paramagnetic particles (“ferrofluid” [FF]). FF-loaded endosomes can be purified from cell homogenates by a magnet (Figure 1A).36,37 Using antibodies against known regulators of endosome maturation, immunoblotting was used to determine to which compartment of the endocytic continuum FF particles were transported in untreated or manipulated cells. If a manipulation blocked LE-LYS fusion, FF would not be transported to LYS, and FF endosomes would contain low levels of LYS-specific proteins (Figure 1B, 4). Lower concentrations of a protein of interest (POI) on FF endosomes could also be due to decreased recruitment of the POI from the cytosol (Figure 1B, 2) or an overall decrease in cellular POI levels (Figure 1B, 3). To distinguish between these scenarios, we have quantified the POI levels in various subcellular fractions (Figure 1B; endosomes, lysate [PNS], membranes, cytosol [cyt]).
Figure 1. Delivery of endocytosed cargo to LEs/LYSs requires Rab7, Arl8a and Arl8b, HOPS, and BORC.

(A) Scheme of pulse/chase labeling and purification of FF endosomes.
(B) Possible scenarios when analyzing WT versus KO FF endosomes. PNS, postnuclear supernatant; cyt, cytosol. See main text for details.
(C–J) FF endosomes purified from HeLa WT or KO cells and PNSs were analyzed for LAMP1 (C–F) or mature cathepsin D (mCathD; G–J) by immunoblotting.
(C) Representative LAMP1 immunoblots of PNSs and FF endosomes.
(D) Quantification of immunoblots as in (C). PNS and LYS samples were normalized to the PNS and LYS sample type with the strongest signal.
(E) Representative LAMP1 immunoblots of PNSs and FF endosomes from HeLa WT or Rab7 KO cells. Eletrophoretic mobility of LAMP1 was altered in PNSs and LYSs from Rab7 KO cells as reported previously.38
(G–J) Done as (C)–(F) but with mCathD antibody. A membrane (mem) fraction was used for whole-cell analysis (G and I).
(K–N) Endosomes purified from WT or KO HeLa cells after a 2 or 24 h chase of FF were analyzed for mCathD.
Data are means and SDs from 3 (D, H, L, and N) or 4 (F and J) independent experiments.
Cargo delivery to LYSs requires Rab7, Arl8a and Arl8b, HOPS, and BORC
To test whether Rab7, Arl8, HOPS, and BORC support encounter and fusion of endosomes and LYSs, we analyzed whether FF was delivered to a compartment containing LAMP1 in wild-type (WT) or knockout (KO) HeLa cells in Rab7, Arl8b, Arl8a and Arl8b, the HOPS subunit Vps41, myrlysin, or diaskedin. As LAMPs are enriched in LYS,39 the presence of LAMP1 in FF endosomes may be used as a measure of LE-LYS fusion. Rab7 KO cells were generated from a different HeLa WT clone and were therefore compared with their own parental WT cells in all experiments.
Cells were incubated with FF for 30 min (pulse), washed to remove extracellular FF, and incubated in medium without FF for 120 min (chase). Post-nuclear cell homogenates (PNSs) and FF endosomes were prepared and analyzed for LAMP1 by immunoblotting. Similar to murine J774E cells,36 endosomes purified after 120 min of chase were strongly enriched in LAMP1 compared with PNSs (Figures 1C and 1E, WT), indicating that FF was delivered to LEs/LYSs within the pulse/chase period. LAMP1 concentrations in endosomes purified from Rab7, Arl8a and Arl8b, Vps41, myrlysin, or diaskedin KO cells were decreased compared with WT compartments (Figures 1C–1F, LYS), albeit total cellular levels of LAMP1 were similar in WT and KO cells (Figures 1C–1F, PNS). Thus, apparently, fusion between FF endosomes and LAMP1-containing LEs/LYSs was inhibited in KO cells.
To test this hypothesis, we analyzed FF endosomes for the presence of mature cathepsin D (mCathD), the bulk of which is in LYSs.39 Endosomes from Rab7, Vps41, Arl8a and Arl8b, myrlysin, or diaskedin KO cells contained much less mCathD than WT compartments (Figures 1G–1J, LYS). Total levels of mCathD were similar in WT and KO cells as revealed by analysis of PNSs (Figures 1G and 1I, PNS) and of membrane fractions prepared by ultracentrifugation of PNSs (Figures 1G and 1I, mem), indicating that KO cells possess normal quantities of mCathD-positive compartments whose fusion with FF endosomes is inhibited. This conclusion is further supported by the fact that after 24 h of chase, endosomes purified from KO cells had acquired similar amounts of mCathD as WT endosomes (Figures 1K–1N), suggesting that LE-LYS fusion is delayed, but not abolished, in KO cells. The only exception were endosomes purified from Arl8b KO cells, which had only a little less LAMP1 and mCathD (Figures 1D and 1H) than WT endosomes, suggesting modest inhibition of LE-LYS fusion in the absence of Arl8b only. This is consistent with Arl8a and Arl8b having at least partially redundant functions.
To test whether reduced fusion with LYSs was the reason that endosomes in KO cells acquired less LAMP1 and mCathD, we analyzed microscopically whether fluorescent dextrans were delivered to acidified compartments in WT and KO cells. Cells were pulsed for 4 h with ATTO488 dextran, non-internalized dextran was removed, and cells were incubated in dextran-free medium for 90 min to allow transport of endocytosed dextran toward LYSs. Cells were then incubated with the acidification reporter LysoTracker Red (LT). Colocalization between dextran-labeled endosomes and LT was decreased in Rab7, Arl8a and Arl8b, Vps41, myrlysin, or diaskedin KO, but not in Arl8b KO, cells (Figures 2A–2H and 2I–2L). KO cells did not accumulate less LT than WT cells as revealed by measuring the LT fluorescence per cell in a fluorometer (Figure S2). Thus, the decreased colocalization of dextrans with LT in KO cells was caused by inhibition of LE-LYS fusion and not by a generalized defect in endosome acidification.
Figure 2. Delivery of endocytosed cargo to acid compartments requires Rab7, Arl8a and Arl8b, HOPS, and BORC.

HeLa WT or KO cells were incubated with ATTO488 dextran and LT.
(A–H) Representative fluorescence micrographs. Cell outlines are indicated. The boxed areas are shown at higher magnification (4.75-fold). Scale bar: 20 μm.
(I–L) Mander’s colocalization coefficients 1 and 2 (M1 and M2) for the overlap of ATTO488 and LT. M1 indicates how many ATTO488 endosomes contain LT, and M2 indicates how many LT endosomes contain ATTO488. M1 and M2 in WT cells were set as 100%.
Data are means and SD from 3 independent experiments.
Endosomes acquire Rab7 and Vps41 before Arl8
Rab7, Arl8a and Arl8b, HOPS, and BORC were required for the transport of endocytosed cargoes to LYSs (Figures 1 and 2), and these factors interact with each other manifold.6,7,20,26 To test in which order these factors are recruited to maturing endosomes, we analyzed endosomes purified from WT HeLa cells at different times after endocytosis of FF for Rab7, Arl8, HOPS, BORC, and LAMP1. Endosomes acquired Rab7 and Vps41/HOPS with similar kinetics (Figures 3A, 3C, and 3D) and before LAMP1, lyspersin/BORC, and Arl8a and Arl8b (Figures 3A, 3E, and 3F). Consistently, Arl8 has been proposed to mark more mature endosomes than Rab735 and to help inactivate and release Rab7 from Arl8-containing compartments, which results in a Rab7-to-Arl8 switch on maturing endosomes.26 Therefore, FF should pass through a Rab7/Arl8-hybrid compartment before ending up in an Arl8-only compartment. Indeed, FF endosomes isolated 2 h after endocytosis contained both, Arl8b-only (Figure 3G, magenta arrowheads), and Arl8b/Rab7-hybrid compartments (Figure 3G, green arrowheads), suggesting that endocytosed cargoes are delivered to terminal Arl8-only compartments via Rab7/Arl8-hybrid endosomes.
Figure 3. Endosomes acquire HOPS and Rab7 before LAMP1, BORC, and Arl8.
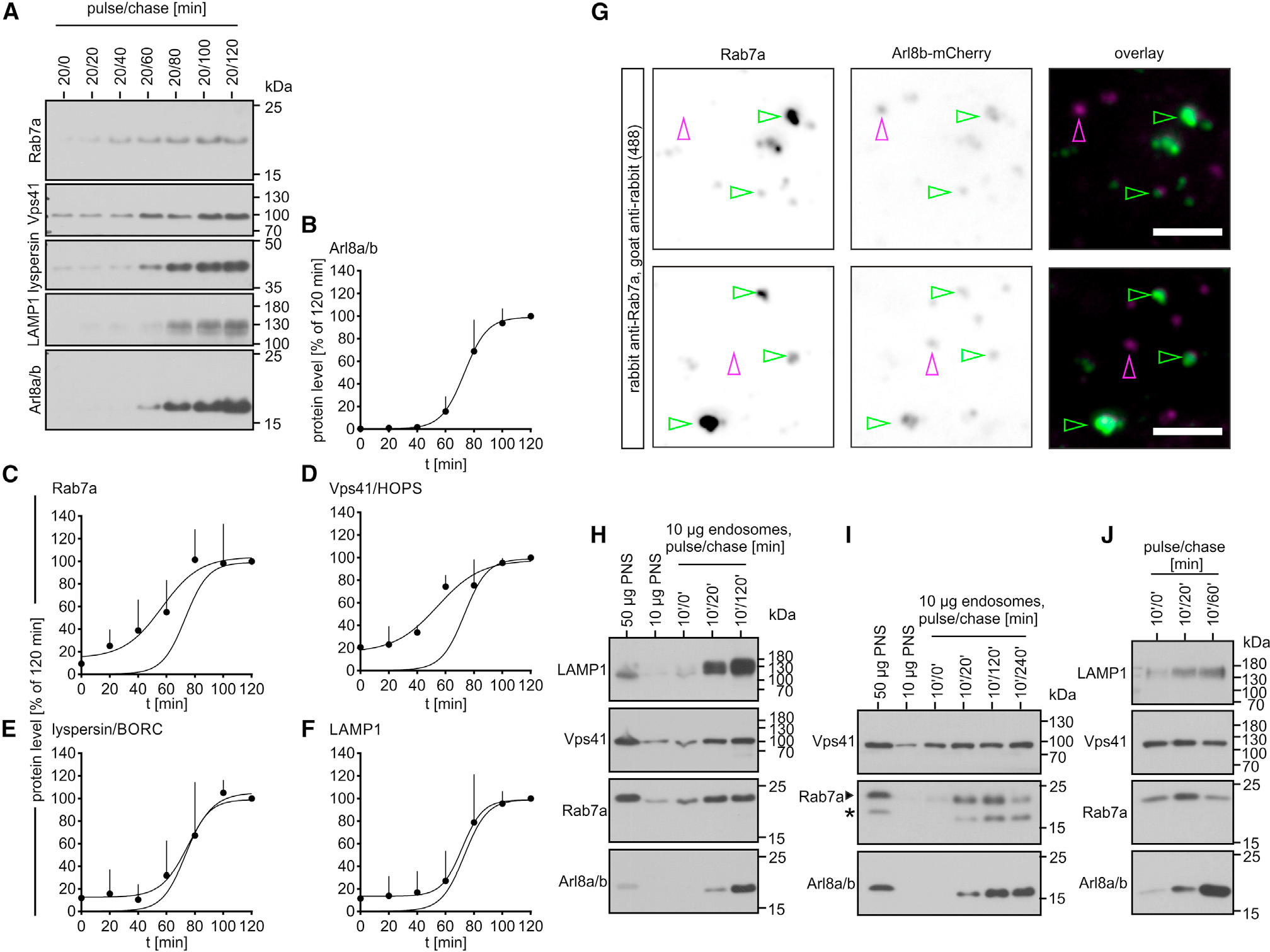
(A–F) FF endosomes were purified from HeLa WT cells after 20 min pulse and the indicated chase periods and analyzed by immunoblotting. (A) Representative immunoblots quantified in (B)–(F). For each protein analyzed, protein levels in 20/120 min endosomes were set as 100%. The curve showing the kinetics of Arl8 acquisition (B) is also shown in (C)–(F). Data are means and SDs from 4 independent experiments.
(G) FF endosomes were purified after 20/120 min (pulse/chase) from HeLa cells expressing Arl8b-mCherry and analyzed for Rab7 by immunofluorescence (IF) microscopy (Rab7 antibody [Abcam]). Representative micrographs. Arrowheads indicate Arl8b-containing endosomes colocalizing with Rab7 (green) or not (magenta). Bar: 2.5 μm.
(H–I) Immunoblot analysis of different-aged FF endosomes from J774E cells. The asterisk in (I) indicates the position of Arl8 as the blot was stained for Arl8 before detection of Rab7.
(J) Different-aged LBPs from J774E cells were analyzed by immunoblotting.
As FF endosomes still contained Rab7 after 120 min of chase (Figures 3A and 3C), we tested whether Rab7 was lost after long chase periods. However, even after 24 h of chase, purified endosomes contained copious amounts of Rab7, whereas the EE markers transferrin receptor (TfR) and EE autoantigen-1 (EEA1) were absent (Figure S3). Interestingly, TfR and EEA1 were retained for more than 6 h of chase even though Arl8 was almost fully present at 4 h of chase (Figure S3). We interpret this observation as follows: non-ingested FF is not entirely removed in the post-pulse wash. Consequently, even after prolonged chase periods, endosome preparations are contaminated by earlier-stage endosomes until the plasma-membrane-bound pool of FF is fully ingested. This caveat still allows us to analyze when endosomes acquire late markers but precludes the analysis of how endosomes exchange early for late and, especially, late for lysosomal markers.
In J774E cells, endosomes lose EE and pick up LE markers within 15–25 min, and they lose LE and accumulate LYS markers within 125 min.36 Apparently, delivery of FF toward LYSs is faster and more synchronous in J774E than in HeLa cells. To test whether endosomes lose Rab7 after recruiting Arl8, we studied the kinetics of endosome maturation in J774E cells. Endosomes acquired late endocytic markers Rab7 and Vps41 before Arl8 (Figure 3H), similar to HeLa cells. Moreover, FF compartments did not lose Rab7 by 120 min (Figure 3H) but lost some of it by 240 min of chase (Figure 3I). Latex bead phagosomes (LBPs) purified from J774E cells, as specialized endosomes, also acquired Vps41 and Rab7 before Arl8 and partially released Rab7 after acquisition of Arl8 (Figure 3J). In sum, our data suggest that endosomes and phagosomes recruit Rab7 and Vps41 before Arl8 and that Rab7-to-Arl8 conversion is a common theme in endosome26 and phagosome maturation.
Rab7 and Vps41 act upstream of Arl8
Because Rab7 and Vps41 are recruited to endosomes before Arl8, they may also act upstream of Arl8. To determine the sequence of action of Rab7, Arl8a, Arl8b, HOPS, and BORC in endosome maturation, we tested whether genetic deletion of any of these factors changes the recruitment of the others to endosomes. To this end, we purified FF endosomes after 120 min of chase from HeLa cells lacking Rab7, Arl8b, Arl8a and Arl8b, Vps41, myrlysin, or diaskedin or from HeLa WT cells. KO of the BORC subunits myrlysin or diaskedin markedly reduced the levels of Arl8 on purified endosomes (Figures 4A and 4C) as expected from the role of BORC in anchoring Arl8 to membranes.14
Figure 4. Rab7 and HOPS are required for the acquisition of Arl8 and BORC.
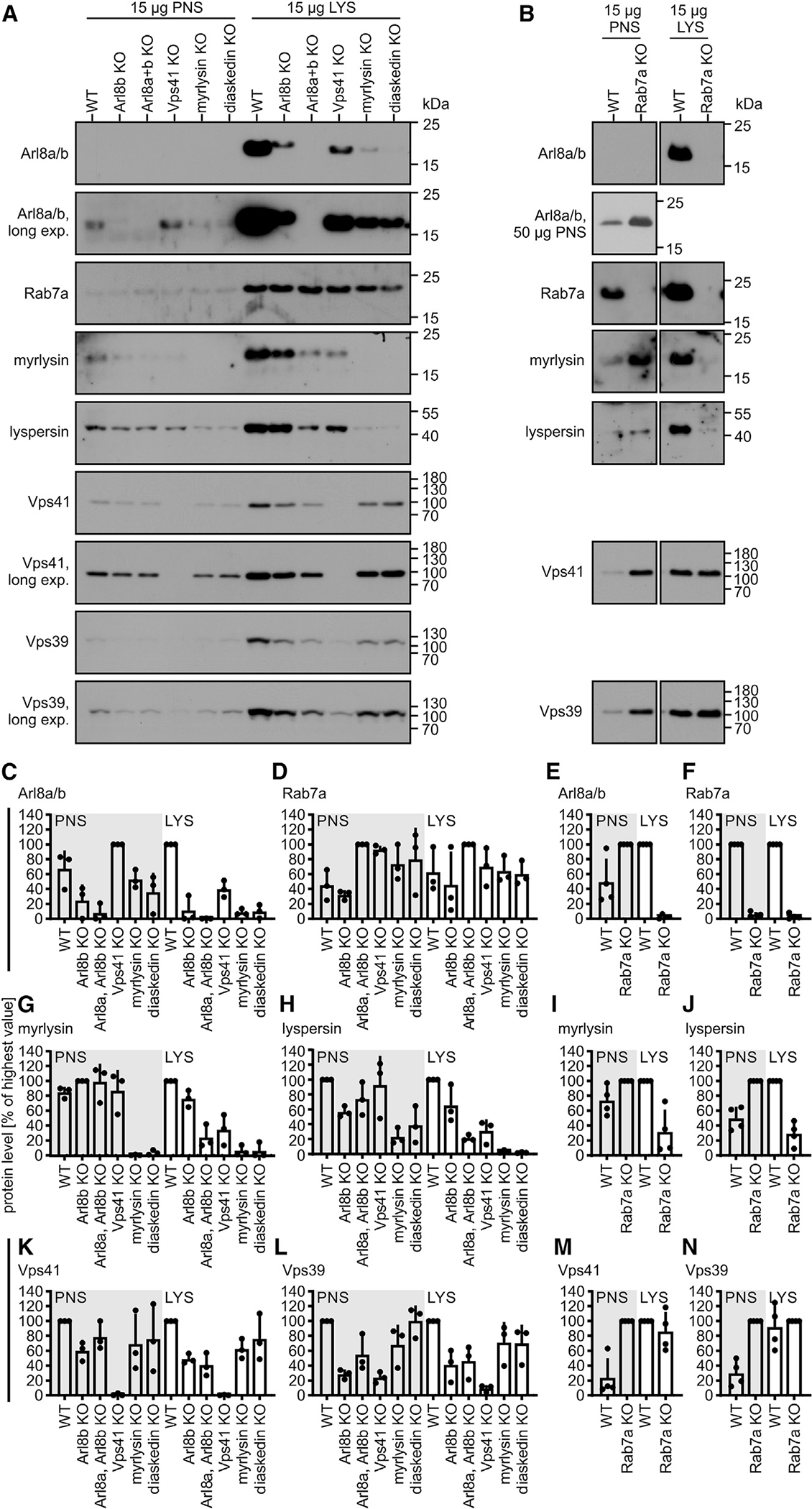
FF endosomes (30/120 min [pulse/chase]) and PNSs from HeLa WT or KO cells were analyzed by immunoblotting.
(A and B) Immunoblots from representative experiments quantified in (C)–(N). PNS and LYS samples were normalized to the PNS and LYS sample with the strongest signal.
(B) Data shown are from identical immunoblots.
Data are means and SDs from 3 (C, D, G, H, K, and L) or 4 (E, F, I, J, M, and N) independent experiments.
Interestingly, endosomes purified from Rab7 or Vps41 KO cells also contained much less Arl8 than WT compartments (Figures 4A–4C and 4E), indicating that Rab7 and Vps41 are not only recruited before but are also required for the acquisition of Arl8. Similarly, in J774E cells, silencing of the expression of Rab7 or Vps41 by small-interfering RNAs (siRNAs) decreased Arl8 acquisition by endosomes (Figure S4), validating our observations with HeLa cells. FF endosomes from Rab7 or Vps41 KO HeLa cells also contained much less of the BORC subunits myrlysin and lyspersin relative to WT endosomes (Figures 4G–4J), indicating that endosomes acquire Arl8 together with its membrane anchor BORC.
The reduced acquisition of Arl8 by endosomes in Rab7 or Vps41 KO cells was neither caused by a decrease of Arl8 levels in these cells (Figures 4C and 4E, PNS) nor by a decrease in recruitment of Arl8 from soluble cytosolic pools (Figures S5A and S5B). Thus, apparently, endosomes acquire Arl8 when Rab7- and HOPS-containing endosomes fuse with downstream Arl8-containing compartments.
To test this hypothesis, we incubated HeLa WT cells with FF for 20/60 min (pulse/chase). By this time, FF endosomes contain about 80% of the amount of Vps41 that they acquire during a 20/120 min period, but they contain little Arl8 (Figure 3D). To test whether acquisition of Arl8 requires membrane fusion, we then added N-ethylmaleimide (NEM), which inhibits SNARE-mediated fusion.40 As a control, cells were incubated with NEM neutralized by an excess of dithiothreitol (DTT). Cells were then incubated for another 60 min at 37°C to allow endosome maturation to complete (Figure S5C). Endosomes from NEM-treated cells contained much less Arl8 than compartments from cells treated with inactivated NEM (Figures S5D–S5F). Notably, treatment of cells with NEM did not release Arl8 or Vps41 into the cytosol (Figure S5G). Thus, endosomes acquire Arl8 as they fuse with preexisting Arl8-containing LYSs.
Rab7 and Vps41 play different roles in endosome maturation
We expected that Rab7 and HOPS would directly drive fusion of FF-containing LEs with LYSs. If so, then Arl8 might not be acquired by endosomes in Rab7 or Vps41 KO cells even after long chase periods. In fact, even after 24 h of chase, FF endosomes lacked Arl8 in Vps41 KO (Figures 5A and 5B) but were Arl8 positive in Rab7 KO cells (Figures 5C and 5D). This indicated that HOPS is indispensable for LE-LYS fusion, whereas Rab7 is not.
Figure 5. HOPS is indispensable for the acquisition of Arl8, but Rab7 is not.

(A–D) FF endosomes were purified after 30 min pulse and a 2 or 24 h chase from Vps41 KO (A and B), Rab7 KO (C and D), or the parental WT cells.
(A and C) Representative immunoblots quantified in (B) and (D). Data were normalized to the Arl8 contents of FF compartments purified from WT cells after 24 h of chase and are means and SDs from 3 independent experiments.
(E and F) HeLa WT or Rab7 KO cells were incubated with LT and Hoechst 33342. Representative fluorescence micrographs. Cell outlines are marked. Scale bars: 20 μm.
(G and H) HeLa WT cells were incubated with 10 μM nocodazole, 100 μM dynarrestin, or DMSO for 30 min at 37°C. FF was added for 30/120 min (pulse/chase) in the presence of the drugs or DMSO. PNSs and FF compartments were analyzed by immunoblotting.
(I) PNSs and FF endosomes from cells expressing GFP or GFP-Rab7 T22N were analyzed by immunoblotting.
Data in (G)–(I) are representative of 2 independent experiments.
Alternatively, Rab7 may contribute to LE-LYS fusion by regulating microtubule- and dynein-dependent transport of endosomes toward perinuclear LYSs.16 In fact, acidic compartments were accumulated around the nucleus in WT cells (Figure 5E) but dispersed throughout the cytoplasm in Rab7 KO cells (Figure 5F). Moreover, endosomes purified from cells treated with the microtubule-destabilizing drug nocodazole or the dynein inhibitor dynarrestin41 or expressing the dominant-negative GDP-locked T22N variant of Rab78 contained less Arl8 than endosomes from mock-treated cells (Figures 5G–5I). Thus, apparently, Rab7-regulated dynein-dependent transport precedes endosome fusion with Arl8-containing LYSs.
Rab2a recruits HOPS to LEs
Endosomes acquired normal amounts of HOPS in Rab7 KO cells (Figures 4M and 4N) and normal amounts of Rab7 in HOPS KO cells (Figure 4D). Thus, Rab7 and HOPS are recruited to maturing endosomes independently of each other. As LEs are free of Arl8, this raises the question of how HOPS is bound to LEs if not through Rab7.
Rab2a was a good candidate as it regulates endosome maturation and binds to the Vps39 subunit of HOPS.27,42,43 To test whether Rab2a recruits HOPS to LEs, we purified FF compartments from mock-treated or Rab2a knockdown (KD) HeLa cells. Rab2a KD endosomes contained less Vps41 and Arl8 than compartments from mock-treated cells (Figures 6A, 6C, and 6D), and KD of Rab2a did not cause Arl8 to dissociate from membranes (Figure 6E), suggesting that Rab2a regulates fusion of LEs with Arl8-containing LYSs by recruiting/anchoring HOPS to LEs.
Figure 6. Rab2a recruits HOPS to LEs.
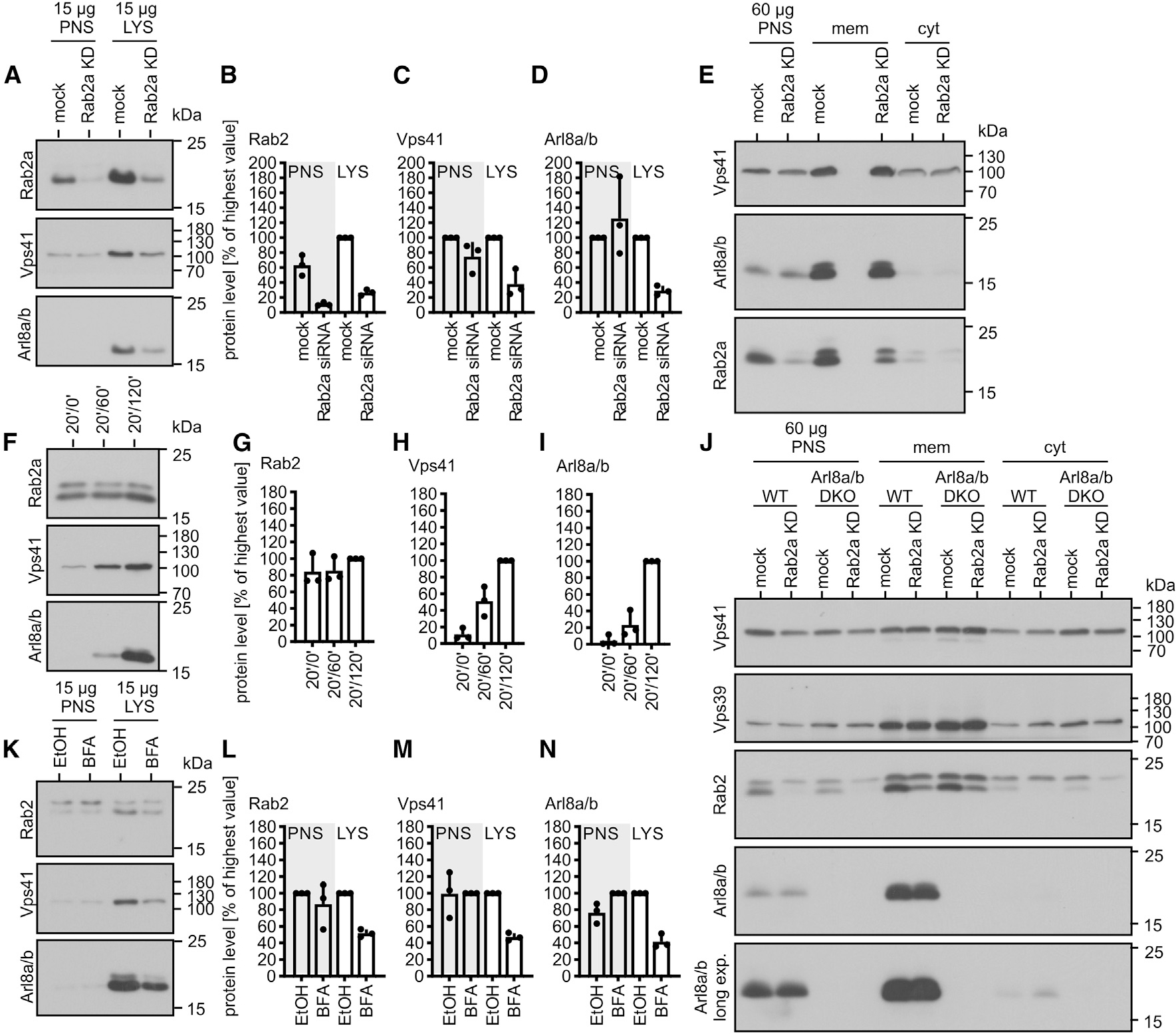
(A–D) PNSs and FF endosomes (20/120 min [pulse/chase]) from mock-treated or Rab2a KD cells were analyzed by immunoblotting. Representative immunoblots quantified in (B)–(D).
(E) PNSs, membrane (mem), and soluble protein (cyt) fractions from mock-treated or Rab2a KD cells were analyzed by immunoblotting.
(F–I) Immunoblot analysis of different-aged FF endosomes from HeLa WT cells; quantification in (G)–(I). Protein levels in 120 min samples were set as 100%.
(J) PNSs from HeLa WT or Arl8a/b KO transfected with non- or Rab2a-targeting siRNAs were fractionated into membrane-bound and cytosolic proteins as in (E).
(K–N) HeLa cells were treated with 1 μg/mL BFA for 30 min at 37°C. Purified FF endosomes (30/120 min [pulse/chase]) were analyzed by immunoblotting. BFA was present throughout the incubations.
(K) Representative immunoblots quantified in (L)–(N).
Data are means and SDs from 3 independent experiments (B–D, G–I, and L–N).
Notably, KD of Rab2a did not cause redistribution of Vps41 to the cytosol (Figure 6E), which would have been expected if Rab2a was the major anchor factor for HOPS. Assuming that HOPS binds to LEs through Rab2a and to LYSs through Arl8, we asked whether KD of Rab2a in Arl8a and Arl8b KO cells would release HOPS to the cytosol. Intriguingly, neither KO of both Arl8 isoforms nor additional KD of Rab2a decreased levels of HOPS on membranes (Figure 6J), suggesting that HOPS has membrane anchors besides Rab2a and Arl8.
Rab2a largely localizes to the Golgi44 and may enter the endocytic pathway membrane bound along the biosynthetic-secretory route. Lower concentrations of HOPS on Rab2a KD endosomes may thus result from their decreased fusion with a Rab2a-containing transport vesicle that feeds HOPS into the endocytic pathway as hypothesized previously.27,45 In fact, brefeldin A (BFA), a fungal metabolite that blocks the transport along the biosynthetic-secretory pathway,46 decreased endosome acquisition of Rab2a, Vps41, and Arl8 (Figures 6K–6N). As BFA does not release Arl8 from LYSs35 and blocks delivery of endocytosed cargoes to LYSs,46 we conclude that endosomes in BFA-treated cells acquire less Arl8, as they fuse less with LYSs.
Previous studies suggested that Rab2a associates with endosomes only transiently,27,42 but Rab2a was not released from HeLa WT endosomes within 120 min of chase (Figures 6F–6I). We therefore tested whether endosomes lose Rab2a at late stages of maturation using J774E cells, in which transport of FF toward LYSs is more synchronous than in HeLa cells (this study). In J774E, Rab2a peaked on LEs and was almost absent from LYSs (Figure S6A). The same was true for LBPs (Figure S6B), confirming that Rab2a binds to maturing endosomes and phagosomes only transiently.
Sites of LYS tethering on LEs contain HOPS and Rab2 but not Rab7
Three observations indicated that HOPS bridges Rab2 on LEs with Arl8 on LYSs: Rab2a (1) recruits HOPS to LEs (Figures 6A and 6C) and (2) associates with maturing endosomes/phagosomes only transiently (Figure S6), and (3) endosomes acquire Arl8 after Rab2a and HOPS (Figures 3A, 3D, and 6F–6I). Therefore, we hypothesized that LEs possess sites at which Rab2a-anchored HOPS captures Arl8(GTP)-decorated LYSs (Figure 7J). These sites may also contain Rab7, but if they did not, this would support our conclusion that Rab7 does not participate in HOPS-dependent tethering. Thus, we probed LEs for Arl8-binding sites and tested whether these sites contained Rab2a and/or Rab7. Specifically, we incubated isolated late phagosomes with purified Arl8b(Q75L). Arl8b has an acetylated methionine and an amphipathic helix at its N terminus, both of which are required for binding of Arl8b to membranes.35 Yet, E. coli does not acetylate Arl8b. Consequently, recombinant Arl8b purified from E. coli is expected to have low affinity for membranes but to still bind to its effectors.7,24 Binding of Arl8b(Q75L) to phagosomes would therefore indicate an interaction with its effector HOPS rather than membrane insertion.
Figure 7. Sites of LYS tethering on LEs contain HOPS and Rab2 but not Rab7.
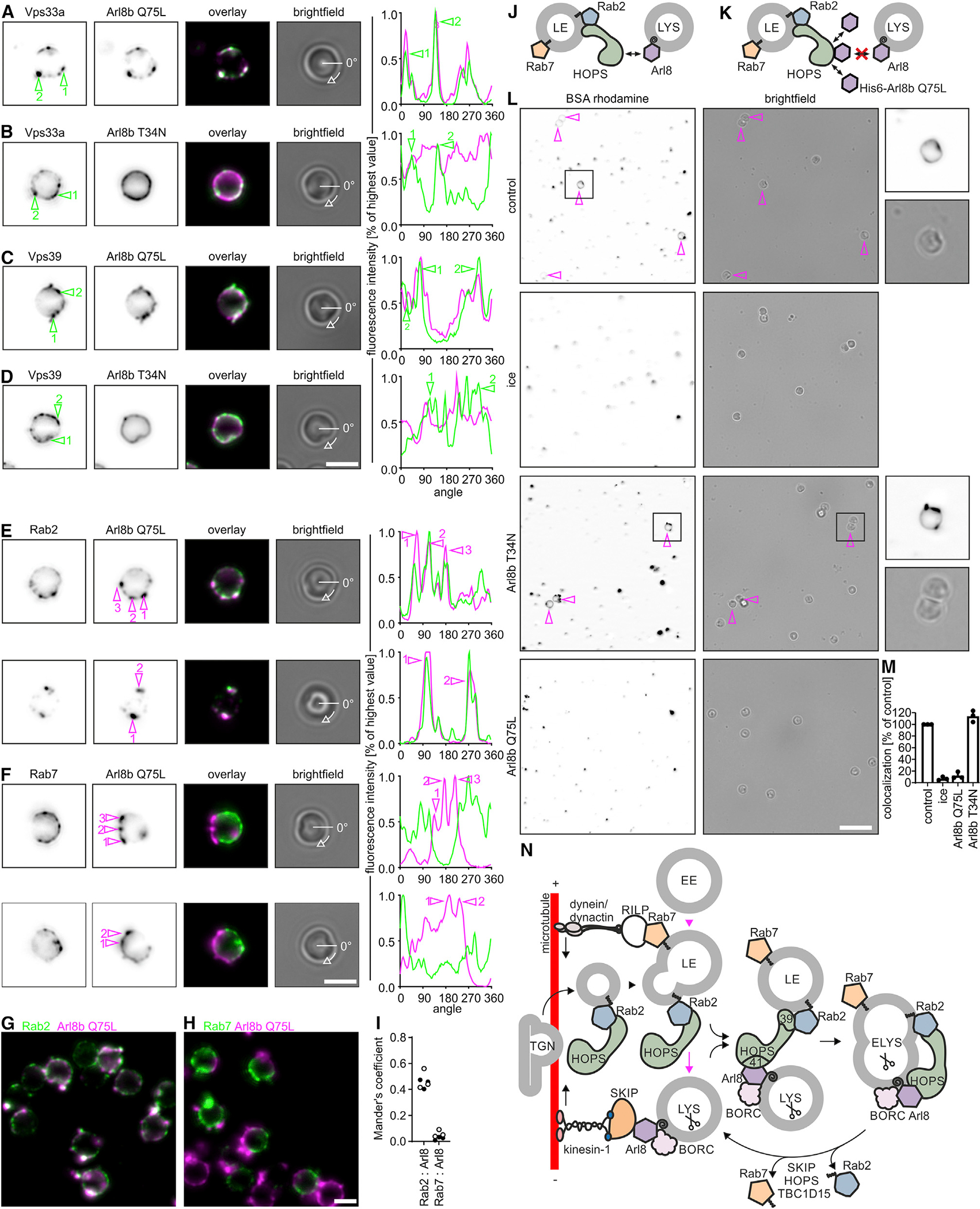
(A–D) Purified late LBPs were incubated with and stained for bound Arl8b(Q75L) (A and C) or Arl8b(T34N) (B and D) and for HOPS (Vps33a, A and B, or Vps39, C and D). Representative fluorescence micrographs. Fluorescence intensities along the circumference of the phagosome for Arl8 or HOPS staining were plotted against the rotation angle, starting at 0 degrees as marked in the bright-field images. Sites of HOPS enrichment are numbered and highlighted in micrographs and fluorescence intensity plots.
(E and F) Similar to (A)–(D), but phagosomes were stained for Arl8b(Q75L) and endogenous Rab2a (E) or Rab7 (F). Sites of Arl8 binding are marked. Data are representative of 2 independent experiments. Bar: 2 μm.
(G and H) Larger sections of representative micrographs from experiments in (E) and (F). Bar: 2 μm.
(I) Mander’s coefficients for the overlap between Rab2a or Rab7 and Arl8b(Q75L). Data points refer to Mander’s coefficients determined for each one micrograph with >10 phagosomes. Black and white data points display results from 2 independent experiments.
(J) Hypothesis how LE-LYS tethering works.
(K) Experimental setup to identify Arl8b(Q75L)-binding sites on LEs as sites of LYS tethering.
(L) Late LBPs were preincubated with purified Arl8b(Q75L) or Arl8b(T34N) and tested for fusion with BSA-rhodamine-loaded LYSs. Representative micrographs. Bar: 10 μm. Magenta arrows indicate LBPs colocalizing with the LYS fluor. Boxed areas are shown at higher magnification.
(M) Colocalization of LBPs with BSA-rhodamine. Data are means ± SD from 3 independent experiments.
(N) Working model of endosome maturation. See main text for details. Scissors indicate the presence of mCathD in ELYSs and LYSs. SNAREs have been omitted for reasons of clarity.
Phagosomes were incubated with Arl8b(Q75L) and analyzed by fluorescence microscopy for its colocalization with HOPS. Arl8b(Q75L) bound to phagosomes in a patchy pattern and colocalized with Vps33a and Vps39 (Figures 7A and 7C), indicating that Arl8b binds to HOPS at sites of LYS tethering. Consistently, GDP-locked Arl8b(T34N), which does not bind to HOPS,7,24 also bound to phagosomes but not specifically to regions of HOPS enrichment (Figures 7B and 7D).
The observed Arl8b(Q75L)-binding sites on phagosomes should contain both HOPS and the GTPase(s) that link HOPS to the LE membrane. Purified late phagosomes were incubated with Arl8b(Q75L) and assayed for its colocalization with endogenous Rab2 or Rab7. Arl8b(Q75L)-positive spots contained Rab2 but not Rab7 (Figures 7E–7I), further supporting our hypotheses that HOPS links Rab2 on late phagosomes/endosomes with Arl8 on LYSs and that Rab7 does not directly participate in HOPS-dependent tethering.
Notably, LBPs preincubated with Arl8b(Q75L) did not fuse with LYSs in a cell-free assay of fusion, suggesting that Arl8b(Q75L)-binding sites on late phagosomes are sites where tethering/fusion with LYSs occurs (Figures 7J–7M). In support of this hypothesis, Arl8b(T34N), which binds to phagosomes in places devoid of HOPS, did not block phagosome-LYS fusion (Figures 7L and 7M). In conclusion, binding of Arl8(Q75L) to late phagosomes/endosomes is a mimic of LYS tethering, during which HOPS and Rab2 on LEs cooperatively capture Arl8(GTP)-decorated LYSs.
DISCUSSION
A biochemical approach to study endosome maturation
Several microscopy studies have reported that the small GTPases Rab7 and Arl8 are indispensable for the delivery of endocytosed cargoes to LYSs.6,7,16,24–26,47 In these studies, POIs were analyzed for colocalization with each other or with marker proteins of EEs or LEs/LYSs using fluorescent fusion proteins. Despite their scientific merit, these studies have not revealed the order in which Rab7, Arl8, and their interactors are recruited to maturing endosomes. Moreover, as proteins that are present in LYSs but absent from LEs have not yet been identified,48 it is unclear whether Rab7, Arl8, and their interactors localize to LEs, to LYSs, or to both.6,26
We set out to analyze the order of recruitment of small GTPases and their interactors by biochemically analyzing endosomes purified after endocytosis of FF, which allows us to simultaneously assess the kinetics of acquisition and the loss of several endocytic marker proteins. This approach analyzes endogenous proteins, which is beneficial as ectopic expression of Rab7, Arl8, or of their effectors can alter the subcellular localization, protein composition, and pH of endosomes.6,7,14,26,49
ELYSs form when Rab7-containing LEs fuse with Arl8-containing LYSs
Our data indicate that endosome maturation comprises a late stage characterized by the presence of Rab7, HOPS and some LAMP1 and a terminal stage specified by Rab7, HOPS, LAMP1, mCathD, BORC, and Arl8. Transition between these stages requires NSF and therefore membrane fusion (this study). As endocytosed cargoes pass through EEs and LEs before they are degraded in ELYSs, the fusion product of LEs and LYSs,1 we propose that our two sequential late endocytic stages represent LEs and ELYSs, respectively.
Endosome maturation likely involves a Rab7-to-Arl8 switch downstream of the Rab5-to-Rab7 switch between EEs and LEs.26 In fact, we show that endocytosed cargoes pass Rab7/Arl8-hybrid endosomes before ending up in Arl8-only compartments. In theory, Rab7/Arl8 hybrids could form when Arl8-containing LYSs recruit Rab7 from soluble cytosolic pools. However, this is unlikely as the Rab7 GEF Mon1/Ccz1 is absent from LYSs50 and as Arl8 counteracts activation and recruitment of Rab7 by attracting the Rab7-GAP TBC1D15.26 Alternatively, Rab7/Arl8 hybrids could form as Rab7-containing LEs recruit soluble Arl8, which is also unlikely as LEs do not contain the Arl8 membrane receptor BORC and cytosolic Arl8 levels are extremely low (this study). In sum, we conclude that Rab7/Arl8-hybrid compartments are formed by fusion of Rab7-only LEs with Arl8-only LYSs.
This fusion event depends on HOPS and its putative membrane anchors Rab7,22,51 Arl8,7,24 and BORC20 because endocytosed cargo was not transported to LYSs and accumulated in Rab7-only LEs in cells lacking Arl8a and Arl8b, HOPS, or BORC (this study), in line with previous reports.7,20–22,51 Since in Rab7 KD cells, endocytosed cargoes are not transported to LYSs and accumulate in multivesicular LE-like compartments,52 FF endosomes from Rab7 KO cells are likely also LEs despite the absence of Rab7.
Arl8-only compartments may represent terminal storage LYSs. In the current view, terminal storage LYSs are pH neutral, hydrolase-inactive compartments for the storage of acid hydrolases,1 which may correspond to the peripherally localized pH-neutral Arl8-positive compartments containing little Rab7 described by Johnson et al.49 Endocytosed cargoes may be passively delivered from cargo-laden ELYSs to terminal storage LYSs by LYS reformation. During LYS reformation, tubules and vesicles bud off from ELYSs and develop into terminal storage LYSs.1
LYS reformation may be similar to the process of phagosome resolution, during which LYSs are reformed from phagolysosomes.53 Apparently, Rab7 and Arl8 are spatially segregated on Rab7/Arl8-hybrid phagolysosomes before Arl8-only vesicles/tubules bud off and mature into LYSs.53 Likewise, Rab7 and Arl8 are segregated on LEs/LYSs as revealed by stimulated emission depletion microscopy.6 Spatial segregation of Rab7 and Arl8 on phago- and endolysosomes may be achieved as Arl8 attracts Rab7-GAP TBC1D15, which releases Rab7 from Arl8-enriched membrane microdomains.26 It remains to be tested whether Rab7-to-Arl8 conversion in the endocytic pathway is accompanied by fission of Rab7/Arl8-hybrid ELYSs into Rab7-only LEs and Arl8-only LYSs.
Arl8 is a marker of LYSs
LEs and LYSs differ in pH, hydrolase activity, buoyancy, and ultrastructure,5 yet, at the molecular level, distinction between LEs and LYSs is difficult because no proteins have been described for LYSs that do not also occur in LEs.48 Previous studies have used LAMPs and mature cathepsins as LYS markers because the bulk of these proteins are present in LYSs. However, as LAMPs and cathepsins are delivered to LYSs via EEs and/or LEs,39 these proteins will always be also present in pre-LYS compartments. Our data indicate that Arl8 is present in LYSs but absent from LEs, which is consistent with the previous observation that Arl8 does not colocalize with the LE marker mannose 6-phosphate receptor.35 The difference in how endosomes acquire mature cathepsins, and Arl8 became obvious when we analyzed the delivery of Arl8 and mCathD to FF endosomes during a prolonged chase period in Vps41 KO cells. Endosomes purified after 24 h of chase did not contain Arl8 but normal amounts of mCathD, which we interpret as follows: in Vps41 KO cells, FF accumulates in Rab7-positive LEs, which do not fuse with LYSs and therefore do not acquire LYS-specific Arl8. However, these fusion-deficient LEs continue to receive cargo from the biosynthetic route, such as autocatalytic procathepsin D,54 leading over time to accumulation of mCathD in aberrant LEs. Different as in the case with mature cathepsins and LAMPs, Arl8 is not by default present in pre-LYS compartments and thus can be used as a handy tool to distinguish LEs from LYSs and likely also late phagosomes from phagolysosomes or amphisomes from autolysosomes in situ.
Endosome acquisition of HOPS depends on Rab2a
Surprisingly, our data indicate that the recruitment of HOPS to LEs does not depend on Rab7.21,22 Aside from Rab7 and Arl8, previous studies have identified Rab2a,27,42 BORC,20 Plekhm1,6,25 and SKIP7 as potential membrane receptors of HOPS. However, it is likely that BORC does not contribute to recruitment of HOPS to LEs because LEs do not contain Arl8 nor BORC (this study). Plekhm1 and SKIP also do not likely recruit HOPS to LEs because Plekhm16 and presumably also SKIP26 do not bind to HOPS in the absence of Arl8.
By contrast, Rab2a does not depend on a LYS-localized factor to bind to HOPS and thus was a good candidate membrane receptor for HOPS on LEs. Rab2a directly binds to the Vps39 subunit of HOPS27 and, when overexpressed as a GTP-locked variant, recruits Vps39 to Rab7-containing compartments.42,43 Moreover, KD of Rab2 leads to the formation of enlarged Rab7-containing endosomes, which do not fuse with cathepsin L-containing LEs/LYSs.27 However, it has not been tested whether KD or KO of Rab2 decreases association of HOPS with endosomes.
We now show that endosomes from Rab2a KD cells contain less HOPS and less Arl8, which may indicate that Rab2a recruits HOPS to LEs and thus promotes LE fusion with Arl8-containing LYSs. Moreover, our observation that Rab2a localizes to LEs and Arl8 to LYSs fits with a scenario in which HOPS promotes LE-LYS fusion by bridging late endosomal Rab2a and lysosomal Arl8. The fact that Rab2 and HOPS colocalize at sites of LYS tethering on late endocytic compartments (this study) supports this conclusion.
Notably, HOPS localized to cell membranes even in the absence of Rab2a and both isoforms of Arl8. We thus propose that Rab2a and Arl8 strongly promote recruitment of HOPS to LEs and LYSs and that HOPS, once recruited, is held in place by its multiple interactors including RILP, Plekhm1, SKIP, SNAREs, and possibly PIPs.33,34 Still, membrane-bound HOPS seems to require Rab2a and Arl8 to promote fusion because LE-LYS fusion is inhibited in Rab2a KD and Arl8a and Arl8b KO cells (this study).
Rab7 in cargo delivery to LYS: Motility promoter or fusion factor?
We here confirm an earlier report that Rab7 promotes LE-LYS encounter through dynein- and microtubule-dependent transport of LEs toward LYSs.16 Intriguingly, however, Rab7 is dispensable for the recruitment of HOPS to LEs and for LE-LYS fusion, and it is absent from sites of LE-LYS tethering (this study). Thus, Rab7 does not appear to act through HOPS. Yet, Rab7 likely stimulates LE-LYS fusion by means other than transport because fusion between phagosomes, i.e., specialized endosomes, and LYSs reconstituted in vitro does not require microtubules but Rab7.33,36
Working model
Our data and previous work by others fit well with the following working model (Figure 7N): endocytosed cargo passes through EEs and is delivered to LEs containing Rab7. A Rab2a- and HOPS-containing transport vesicle originating from the TGN fuses with and delivers Rab2a and HOPS to LEs. Rab7 is dispensable for the recruitment of HOPS to LEs yet may stabilize its membrane association. Rab7 attracts the dynactin-dynein motor protein complex and moves LEs toward perinuclear Arl8-containing LYS, which promotes LE-LYS encounter. Rab2a-associated HOPS on LEs binds to lysosomal Arl8 and associated BORC forming a tethering bridge and promoting LE-LYS fusion. Alternatively, HOPS associated with Arl8 and BORC on LYSs may contact free Rab2a on LEs.
Once tethering has been achieved, HOPS is proposed to promote formation of trans-complexes between the SNARE proteins Syntaxin 7 (Qa), Vti1b (Qb), and Syntaxin 8 (Qc)55 and either of the R-SNAREs Vamp8, Vamp7, or Ykt6,56 leading to LE-LYS fusion. The resulting ELYSs contain Rab7 and Arl8 and release Rab7 through an Arl8-attracted ternary complex of SKIP, HOPS, and the Rab7-GAP TBC1D15, possibly in the course of LYS reformation.
Plekhm1 and SKIP may contribute to HOPS-dependent tethering. Likely, Plekhm1 supports formation of Rab7-Arl8 hybrid ELYSs, whereas SKIP removes Rab7 from these hybrids. Hence, the Rab7-to-Arl8 switch between LEs and LYSs (this study and Jongsma et al.26) may be accompanied by a Plekhm1-to-SKIP switch.
The topology of the tethering machinery that we present here only applies to heterotypic LE-LYS fusion. Homotypic fusion of LEs similarly depends on HOPS57 but may use a different combination of HOPS membrane anchors, as LEs do not contain Arl8 (this study and Hofmann and Munro35).
In summary, our study characterizes the order of action of HOPS and its interacting proteins and reveals the topology of the machinery that drives LE-LYS tethering and fusion in mammalian cells.
Limitations of the study
In cells depleted of HOPS or of its interactors, endocytosed cargoes are not delivered to LYSs, which we and others interpret as a defect in LE-LYS fusion. However, strictly speaking, our and previously reported assays did not distinguish whether reduced cargo delivery to LYSs is caused by inhibition of endosome-toward-LYS transport or of endosome-with-LYS fusion. Future studies, using biochemically reconstituted systems such as the one presented here (Figures 7J–7M), will have to determine whether Arl8, Rab7, and their interactors are fusion regulators in their own right or rather transport regulators.
As noted in the results, LYSs prepared from HeLa cells using our FF pulse/chase labeling protocol are contaminated with earlier endocytic compartments (Figure S3).
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andreas Jeschke (jeschke@uni-bonn.de).
Materials availability
New unique materials generated in this study are available upon request.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
J774E macrophage-like cells (mouse, sex: female)60 were cultivated in DMEM containing 5% (v:v) fetal bovine serum (FBS), 1% (v:v) glutamax and 1% (v:v) PenStrep.
HeLa cells (human, sex: female) were cultivated in DMEM containing 5% (v:v) FBS, 1% (v:v) glutamax, 1 mM sodium pyruvate and 1% (v:v) PenStrep at 37°C in a humid atmosphere at 5% CO2. For simplicity, cell culture media for HeLa and J774E cells are referred to as DMEM/FBS, cell culture media without FBS as DMEM/-FBS throughout the manuscript.
METHOD DETAILS
Purification of Arl8b Q75L and Arl8b T34N
Fifty mL of LB broth containing 100 μg/mL ampicillin (LB/amp) were inoculated with E. coli BL21 (DE3) transformed with pET45b(+)-Arl8b Q75L or pET45b(+)-Arl8b T34N and incubated at 37°C for 16 h. Ten mL of the resulting culture were used to inoculate 500 mL of LB/amp. Cultures were incubated at 37°C for 3 h, isopropyl-β-D-thio-galactopyranoside (IPTG) was added to a final concentration of 0.25 mM and cultures were incubated at 26°C for 16 h. Bacteria were harvested, resuspended in lysis buffer (20 mM HEPES, 300 mM KCl, 1.5 mM MgCl2, 10 mM imidazole, pH 7.2 [HCl]) containing 1x protease inhibitor cocktail (PIC; a 50x stock solution contained 5 μg/mL leupeptin, 25 mM 1,10-phenantroline, 25 μg/mL pepstatin A, 5 mM pefabloc) and lysed by sonication. Lysates were centrifuged (16,100 × g, 30 min, 4°C) and supernatants were incubated for 60 min at 4°C with each 1 mL Ni-NTA agarose (Macherey & Nagel) equilibrated in lysis buffer. Ni-NTA beads washed twice in each 20 mL lysis buffer and once in 20 mL wash buffer (20 mM HEPES, 300 mM KCl, 1.5 mM MgCl2, 20 mM imidazole, pH 7.2 [HCl]; 1,800 × g, 2 min, 4°C). To elute proteins, Ni-NTA agarose was washed 3 times with each 1 mL of elution buffer (20 mM HEPES, 300 mM KCl, 1.5 mM MgCl2, 250 mM imidazole, pH 7.2 [HCl]). Eluted proteins were dialyzed against 4 L of HB/1x salts (20 mM HEPES, 0.5 mM EGTA; 100 mM KCl, 1.5 mM MgCl2, pH 7.2 [KOH]) for 2 h at ambient temperature using a nitrocellulose membrane with a 3 kDa cutoff. Proteins were snap-frozen in liquid nitrogen and stored at −80°C.
Generation of HeLa Rab7a KO cells
The Rab7a gene was inactivated in HeLa cells using the CRISPR/Cas9 system. SgRNA (UGAAUUUCUUAUUCACAUAC) (Synthego) was complexed with purified Streptococcus pyogenes Cas9 (Synthego) and delivered into HeLa cells by electroporation (Neon electroporation system, Thermo Fisher). After transfection, cells were transferred to 96 well plates and KO clones were selected by limiting dilution cloning. KO clones were identified by immunoblotting.
Purification of FF-containing compartments
FF-containing compartments were purified from cell cultures as described previously63 with modifications. The following instructions refer to one cell culture dish (10 cm diameter) of HeLa or J774E cells. A suspension of 10 μL FF in 2.5 mL of DMEM/-FBS was sonicated for 30 s, sterile-filtered using a 0.2 μm pore-size filter and warmed to 37°C. Cells were washed once with 3 mL PBS and 2.5 mL of the FF suspension were added. Cells were incubated for 30 min at 37°C (pulse), washed twice with 3 mL PBS, DMEM/FBS was added and cells were incubated at 37°C for 120 min (chase) unless stated otherwise. DMEM/FBS was discarded, 2 mL of cold PBS were added and cells were detached from dishes using a cell scraper. Cell suspensions were centrifuged (440 × g, 4°C, 5 min), the supernatant was discarded and cells were resuspended in 2 mL PBS/EDTA (PBS, 5 mM EDTA [ethylene diamine tetraacetic acid]). Cells were sedimented (440 × g, 4°C, 5 min), the supernatant was discarded and cells were resuspended in 2 mL homogenization buffer (HB, 250 mM sucrose, 20 mM HEPES, 0.5 mM EGTA, pH 7.2). Cell suspensions were centrifuged (440 × g, 4°C, 5 min) and cells were resuspended in 1 mL of HB containing 1x PIC. Cells were broken by 20 strokes in a dounce homogenizer and homogenates were centrifuged (800 × g, 4°C, 5 min) to prepare a postnuclear supernatant (PNS). The PNS was collected, the corresponding pellet was resuspended in HB and centrifuged again (800 × g, 4°C, 1 min). The resulting supernatant was mixed with the PNS and incubated on a DynaMag-2 magnet (Invitrogen, 12321D) for ≥30 min at 4°C. Supernatants were removed and FF compartments were sedimented by centrifugation (450 × g, 5 s). Protein concentrations in the PNS and the FF compartment preparation were determined using Biorad protein assay reagent (Biorad, 5000006) according to the manufacturer’s instructions and using bovine serum albumin (BSA) as a protein standard. PNSs and FF compartment preparations were adjusted to identical protein content and to 1x SDS sample buffer, boiled (95°C, 3 min) and analyzed by SDS-PAGE and immunoblotting. For all compartment preparations in this study at least 4 cell culture dishes (10 cm diameter) of HeLa or J774E cells grown to 80% confluency were used.
Preparation of LBPs from J774E cells
Carboxylate-modified 1 μm latex beads (Polysciences) were coated with BSA,33 using 0.3 mg of BSA for the coating of 4.6 × 1010 particles in 1 mL of MES (2-(N-morpholino)ethanesulfonic acid) buffer (50 mM MES/NaOH, pH 6.8). LBPs were purified from J774E cells as described previously.63 LBP preparations of different maturation stages were obtained using different pulse/chase protocols: Early phagosomes were prepared after 10 min of pulse, late phagosomes after 10 min/20 min (pulse/chase) and phagolysosomes after 30 min/60 min.63 For the preparation of early phagosomes and late phagosomes 20 dishes (10 cm diameter) of J774E macrophages were used, for phagolysosomes 10 dishes. The following protocol refers to one cell culture dish. Cells were washed once with 5 mL PBS and 2.5 mL of a latex bead suspension (32 μL beads per 2.5 mL DMEM/-FBS) were added. After the pulse period, cells were washed thrice with 5 mL PBS and either harvested (early phagosomes) or 3 mL DMEM/FBS (prewarmed to 37°C) were added and cells were further incubated at 37°C for 20 min (late phagosomes) or 60 min (phagolysosomes). After completion of the pulse/chase periods, DMEM/FBS was discarded, 2 mL of cold PBS were added and cells were detached from dishes using a cell scraper. Cell suspensions of 10 or 20 dishes were combined and centrifuged (440 × g, 4°C, 5 min), the supernatant was discarded and cells were resuspended in 20 mL PBS/EDTA. Cells were sedimented (440 × g, 4°C, 5 min), the supernatant was discarded and cells were resuspended in 20 mL HB. Cell suspensions were centrifuged (440 × g, 4°C, 5 min) and cells were resuspended in 2 mL of HB containing 1x PIC. Cells were broken by 20 strokes in a dounce homogenizer and homogenates were centrifuged (800 × g, 4°C, 5 min) to prepare a PNS. The PNS was collected, the corresponding pellet was resuspended in 1 mL HB and centrifuged again (800 × g, 4°C, 1 min). The resulting supernatant was collected and mixed with the PNS. The combined supernatants were transferred to an ultracentrifugation tube and mixed carefully with 3 mL of a 62% sucrose solution (62% [w:v] sucrose, 20 mM HEPES, 0.5 mM EGTA, pH 7.2 [KOH]). The resulting suspension was overlaid with 3 mL of a 25% sucrose solution (25% [w:v] sucrose, 20 mM HEPES, 0.5 mM EGTA, pH 7.2 [KOH]) and 3 mL of HB. The resulting sucrose step gradient was centrifuged in a Beckman Coulter Optima L-80 XP ultracentrifuge (SW40 Ti rotor, 18,300 rpm, 4°C, 30 min) and LBPs were harvested from the 25% sucrose-HB interface. LBP suspensions were diluted 1:1 (per volume) by addition of 20 mM HEPES, 0.5 mM EGTA (pH 7.2, KOH) and sedimented in a Beckman Coulter Optima Max XP table top ultracentrifuge (TLA-55 rotor, 186,000 × g, 30 min, 4°C). LBPs were resuspended in 20 μL of HB. LBP suspensions were adjusted to identical OD600 (i.e., identical LBP numbers64) and to 1x SDS sample buffer, boiled for 5 min at 95°C and analyzed by SDS PAGE and immunoblotting.
Binding of purified Arl8b to phagosomes
Late phagosomes were purified from J774E cells 10 min/20 min (pulse/chase) after phagocytosis of 2 μm latex beads as described for LBPs containing 1 μm latex beads above. Purified LBPs were incubated for 30 min at 37°C with 10 μM hexahistidine-tagged Arl8b(Q75L) or Arl8b(T34N), in HB containing an ATP-regenerating system, 1x salts, and 1 mM DTT.33 LBPs were isolated from reaction mixtures by density gradient centrifugation and fixed onto glass coverslips. Samples were sequentially incubated with PBS/50 mM NH4Cl and with blocking buffer (PBS/4% goat serum) for each 15 min at ambient temperature. LBPs were incubated with a mouse anti-Histidine tag antibody (1:50 in blocking buffer) for 60 min, washed five times with PBS, and incubated with an Alexa 535-conjugated goat anti-mouse antibody (1:200, blocking buffer). Samples were washed five times with PBS and incubated with rabbit anti-Rab2 or rabbit anti-Rab7 (Abcam) antibodies (each 1:50, blocking buffer) for 60 min at ambient temperature. Samples were washed five times with PBS and incubated with an Alexa 488-labeled goat anti-rabbit antibody (30 min, 1:200 in blocking buffer). Samples were washed four times with PBS, twice with ddH2O and mounted in Mowiol. Samples were analyzed using a Zeiss Axio Imager equipped with Apotome and a 100x oil immersion lens.
Cell-free phagosome-lysosome fusion
Cell-free phagosome-lysosome fusion was essentially performed as previously described.33 In brief, late phagosomes purified from J774E cells 10 min/20 min (pulse/chase) after phagocytosis of 2 μm latex beads were incubated for 10 min at 37°C with 20 μM hexahistidine-tagged Arl8b(Q75L) or Arl8b(T34N), in HB containing 1x ATP-regenerating system, 1x salts, and 1 mM DTT. J774E cytosol36 and BSA rhodamine-loaded LYS33 were added to final concentrations of 2 and 0.4 mg/mL of protein, respectively, and reactions were incubated for another 60 min at 37°C. Reactions were incubated with 0.2 mg/mL proteinase K on ice for 15 min and PMSF was added to a final concentration of 1 mM. Reactions were adjusted to 210 μL HB, laid on 1 mL 25% sucrose cushions and centrifuged (1,800 × g, 30 min, 4°C). Phagosomes were harvested from the HB/25% sucrose interface, adjusted to 500 μL HB and 2 mg/mL BSA and spun onto glass coverslips in 24 well plates (1,800 × g, 20 min, 4°C). Supernatants were removed and phagosomes were fixed with 4% formaldehyde (FA) in HB for 16 h at 4°C. Samples were mounted in mowiol and LBPs were assayed for colocalization with the lysosomal fluor by fluorescence microscopy.
Analysis of cytosolic and membrane proteins
In some experiments, we determined whether a given protein was membrane-bound or cytosolic. To this end, cells were treated as specified in figure legends, the medium was discarded, 2 mL (per 10 cm dish) of cold PBS were added and cells were scraped off dishes using a cell scraper. Cells were sequentially washed with each 1 mL of PBS/EDTA and of HB (440 × g, 4°C, 5 min) and resuspended in 750 μL HB containing 1x PIC. Cells were homogenized by 20 strokes in a dounce homogenizer and homogenates were centrifuged (800 × g, 4°C, 5 min) to prepare PNSs. PNSs were adjusted to 120 μg of protein and to 1 mL of volume by addition of HB and centrifuged using a Beckman Coulter Optima Max XP table top ultracentrifuge (TLA-55 rotor, 186,000 × g, 4°C, 30 min). Pellets (membrane fraction) were resuspended in 2x SDS sample buffer and boiled (95°C, 3 min). Supernatants (cytosol fraction) were adjusted to 12% (v:v) trichloroacetic acid (TCA) and incubated for 16 h at 4°C to precipitate proteins. Precipitated proteins were sedimented (16,100 × g, 4°C, 30 min) and resuspended in 2x SDS-sample buffer. Samples were neutralized by addition of 1 M Tris base (pH 9.0) and boiled (95°C, 3 min). The membrane and cytosol fractions and 60 μg of the corresponding PNS were analyzed by SDS-PAGE and immunoblotting.
Preparation of ATTO488-dextran
For preparation of ATTO488-dextran, 5 mg/mL 10 kDa amino-dextran in 0.1 M NaHCO3 were mixed with ATTO488 NHS-ester (dissolved in DMSO) at a final concentration of 0.108 mg/mL, incubated for 3 h at ambient temperature and dialyzed against an excess of PBS for 16 h at 4°C.
Dextran delivery to acid compartments
HeLa cells (1 × 105) were seeded onto glass coverslips in 24 well plates one day before the experiment. The medium was displaced, cells were washed once with PBS and incubated with 1.2 mg/mL ATTO488-labelled dextran in DMEM/FBS for 210 min (pulse). Cells were washed thrice with PBS and DMEM/FBS was added for 90 min (chase). To stain acidic compartments, the medium was replaced by DMEM/FBS containing 100 nM LT and cells were incubated for 30 min at 37°C. Cells were washed thrice with PBS. To disrupt microtubules and cause redistribution of acidic compartments within cells, DMEM/FBS containing 10 μM nocodazole was added for 20 min at 37°C. Coverslips were mounted in 0.5% (w:v) low melting agarose in PBS and cells were analyzed by epifluorescence microscopy using a Zeiss Axio Observer epifluorescence microscope equipped with a 100x oil immersion objective and a HXP lamp.
Analysis of colocalization between ATTO488-dextran and LT was performed using ImageJ.61 In brief, micrographs corresponding to the fluorescence signals detected in the red (LT) or in the green (ATTO488) channel were exported as tif files, opened in ImageJ and transformed to 8-bit. Fluorescence maxima (corresponding to the center of mostly round-shaped ATTO488 and LT compartments, respectively) in both micrographs were detected using the ‘Find Maxima’-command (noise: 10, output: single points) of the ‘Process’-tab and for each micrograph a binary image with points marking the position of the fluorescence maxima was generated. These points were dilated thrice using the ‘dilate’-command (Process>Binary>Dilate) which adds pixels to the edge of black objects. Using the JACoP-Plugin,62 the Mander’s correlation coefficients (M1 and M2) for images displaying fluorescence maxima of either LT or ATTO488 vesicles as single, uniform, same-size, same-intensity points were determined. Accidental overlap between fluorescence maxima was rare, as assessed by determining M1 and M2 for pairs of images in which one had been flipped vertically (data not shown). Determining the overlap between the fluorescence maxima distinguishes between ATTO488-containing endosomes being attached or close to LT-positive compartments and ATTO488-compartments that contain LT, similar to the distance-based endosome content-mixing readout used in.65
Fluorometric analysis of LT staining
Cells (1 × 104) were seeded in 96-well plates and cultured for 16 h before the experiment. Cells were washed once with PBS and DMEM/FBS containing 1 μM of the nucleic acid stain Syto13 and 100 nM LT was added for 30 min at 37°C. Cells were washed thrice with PBS and analyzed for Syto13 and LT fluorescence in a Biotek FLx800 fluorescence microplate reader at 485 (±10) nm excitation and 516 (±10) nm emission for Syto13 and at 560 (±20) nm excitation and 625 (±16) nm emission for LT. To normalize the LT fluorescence to the number of cells analyzed per well, the ratio between LT fluorescence and Syto13 fluorescence was determined.
Transfection of cells with siRNAs
Cells were sub-cultivated at a 1:3 ratio 1 day before transfection. The following instructions refer to one cell culture dish of HeLa cells. Cells were washed once with 3 mL PBS and detached by incubation with 1 mL of 0.25% trypsin-EDTA (Gibco, 25200056). Two mL DMEM/FBS were added and detached cells were resuspended. Cells were sedimented (440 × g, 5 min) and washed twice in 3 mL PBS (440 × g, 5 min). Cells were counted and the number of cells required for transfection was sedimented (440 × g, 5 min). Cells were resuspended in Optimem (ambient temperature) at 4 × 106 cells per 100 μL. One μL of a 40 μM stock of siRNAs was transferred to a 4 mm electroporation cuvette and 100 μL of cells suspended in Optimem were added, resulting in a final siRNA concentration of 400 nM. After 3 min at ambient temperature, cells were electroporated in a Biorad Gene Pulser Xcell electroporator using the following settings: square wave protocol, 0.5 ms pulse length, 1000 V, 2 pulses, 5 s interval, 4 mm cuvette. Cell suspensions were transferred to 10 mL of DMEM/FBS prewarmed to 37°C in a 10 cm cell culture dish.
Transfection of J774E cells was done similarly. Modifications were that cells were washed twice with PBS (3 mL per 10 cm dish) before being harvested from cell culture dishes using a cell scraper and that 8 × 106 of siRNA-transfected cells were seeded per 10 cm cell culture dish. Cells were used for experiments 24 h after transfection. For preparation and immunoblot analysis of FF compartments from siRNA-transfected cells, we routinely used 4 dishes with each 4 × 106 HeLa or 8 × 106 J774E cells.
Cloning of pmCherry-N1-Arl8b
Human Arl8b was amplified from pGEX4T3-hArl8b using the primers listed in the key resources table and ligated into pmCherry-N1 using the XhoI and SacII restriction sites.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse anti-LAMPI (H4A3) | Santa Cruz Biotechnology | Cat#sc-20011; RRID:AB_626853 |
| Rat anti-LAMPI (1D4B) | Santa Cruz Biotechnology | Cat#sc-19992; RRID:AB_2134495 |
| Mouse anti-cathepsin D (C-5) | Santa Cruz Biotechnology | Cat#sc-377124 |
| Mouse anti-Vps41 (D-12) | Santa Cruz Biotechnology | Cat#sc-377118; RRID:AB_2687987 |
| Mouse anti-Arl8a/b (H-8) | Santa Cruz Biotechnology | Cat#sc-398635 |
| Mouse anti-Vps39 (C-5) | Santa Cruz Biotechnology | Cat#sc-514762; RRID:AB_2687985 |
| Mouse anti-CD71 (H68.4) | Santa Cruz Biotechnology | Cat#sc-65882; RRID:AB_1120670 |
| Rabbit anti-cathepsin D | Sigma Aldrich | Cat#219361; RRID:AB_10683361 |
| Rabbit anti-lyspersin | Abcam | Cat#ab247064 |
| Rabbit anti-Rab7a | Abcam | Cat#ab137029; RRID:AB_2629474 |
| Rabbit anti-myrlysin | Proteintech | Cat#17169-1-AP; RRID:AB_2137150 |
| Rabbit anti-Rab2 | Proteintech | Cat#15420-1-AP; RRID:AB_2176874 |
| Rabbit anti-Arl8b | Proteintech | Cat#13049-1-AP; RRID:AB_2059000 |
| Rabbit anti-Vps39 | Proteintech | Cat#16219-1-AP; RRID:AB_11043179 |
| Rabbit anti-Vps33a | Proteintech | Cat#16896-1-AP; RRID:AB_2214916 |
| Rabbit anti-EEA1 | Thermo Scientific | Cat#MA5-14794; RRID:AB_10985824 |
| Mouse anti-Histidine tag | Biorad | Cat#MCA 1396; RRID:AB_322084 |
| Rabbit anti-Rab7a | T. Watts (University of Toronto, Toronto, ON, Canada), Bertram et al.58 | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Ferrofluid (EMG508) | Ferrotec | N/A |
| Polybead Carboxylate 1.0 micron microspheres | Polysciences | Cat#08226-15 |
| Polybead Carboxylate 2.0 micron microspheres | Polysciences | Cat#18327 |
| ATTO488 NHS-ester | ATTO-TEC | Cat#AD488-35 |
| Brefeldin A | Santa Cruz Biotechnology | Cat#sc-200861 |
| N-ethylmaleimide | Sigma Aldrich | Cat#E3876 |
| Nocodazole | Sigma Aldrich | Cat#M1404 |
| Dynarrestin | Sigma Aldrich | Cat#SML2332 |
| Hoechst 33342 | Immunochemistry technologies LLC | Cat#639 |
| Amino dextran 10 kDa | Invitrogen | Cat#D1860 |
| Syto13 | Invitrogen | Cat#S7575 |
| Lysotracker Red DND-99 | Invitrogen | Cat#L7528 |
| Opti-mem I Reduced Serum Medium | Gibco | Cat#31985070 |
| Dulbecco’s modified Eagle medium (DMEM) | Gibco | Cat#31053-028 |
| Sodium pyruvate | Gibco | Cat#11360-070 |
| Penicillin-Streptomycin 10000 U/mL (PenStrep) | Gibco | Cat#15140-122 |
| Fetal bovine serum Standard (FBS) | Pan Biotech | Cat#P30-3306 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HeLa WT cells | Pu et al.14 | N/A |
| HeLa Arl8b KO cells | Guardia et al.19 | N/A |
| HeLa Arl8a Arl8b KO cells | Keren-Kaplan et al.18 | N/A |
| HeLa Vps41 KO cells | Anderson et al.59 | N/A |
| HeLa myrlysin KO cells | Pu et al.14 | N/A |
| HeLa diaskedin KO cells | Jia et al.20 | N/A |
| HeLa Rab7a KO cells | This paper | N/A |
| J774E macrophage-like cells | P.D. Stahl (Washington University, St. Louis, USA), Fiani et al.60 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Mission siRNA universal negative control #1 | Sigma Aldrich | Cat#SICO01-10NMOL |
| Custom siRNA, human Rab2a (5′–3′): GAAGGAGUCUUUGACAUUA[dT][dT]–3′ |
This paper, sequence derived from Lörincz et al.27 | N/A |
| Custom siRNA, mouse Vps41 (5′–3′): UUGGCUUAUUUAGUUAGCAAA[dT][dT] | This paper | N/A |
| Custom siRNA, mouse Rab7a (5′–3′): CCAUCAAACUGGACAAGAA[dT][dT]–3′ | This paper | N/A |
| Primer Arl8b forward (5′–3′) TAAGCACTCGAGATGCTGGCGCTC ATCTCC–3′ |
This paper | N/A |
| Primer Arl8b reverse (5′–3′) TGCTTACCGCGGGCTTCTTCTAGAT TTTGAATGCTGA |
This paper | N/A |
| SgRNA human Rab7a: UGAAUUUCUUAUUCACAUAC |
Synthego | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pET45b(+)-hArl8b T34N | M. Sharma (Department of Biological Sciences, Indian Institute of Science Education and Research | N/A |
| Mohali, Punjab, India), Khatter et al.7 | ||
| pET45b(+)-hArl8b Q75L | M. Sharma, Khatter et al.7 | N/A |
| pGEX4T3-hArl8b | M. Sharma, Khatter et al.7 | N/A |
| pmCherry-N1-hArl8b | This paper | N/A |
| pmCherry-N1 | D. Fürst (Cell Biology Institute, University of Bonn, Germany) | N/A |
| peGFP-C1-Rab7a T22N | B. van Deurs (University of Copenhagen, Copenhagen, Denmark). Bucci et al.8 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | Schneider et al.61 | https://imagej.nih.gov/ij/ |
| Image J plugin JACoP | Bolte and Cordelieres62 |
https://imagej.nih.gov/ij/plugins/
track/jacop.html |
| Image J plugin oval profile | Bill O’Connell |
https://imagej.nih.gov/ij/plugins/
oval-profile.html |
| GraphPad Prism 9 | GraphPad Software |
https://www.graphpad.com/
scientific-software/prism/ |
| MathCad 14 | PTC | N/A |
Transfection of cells with plasmid DNAs
Cells were sub-cultivated the day before transfection, yielding cell cultures grown to approximately 50% of confluency on 10 cm cell culture dishes. Seven hundred μL of 2x HBS (274 mM NaCl, 50 mM HEPES, 11.1 mM glucose, 10 mM KCl, 1.9 mM Na2HPO4, pH 7.05 [NaOH]) were dropwise added to 700 μL of a solution containing 311 mM CaCl2 and 12 μg pmCherry-N1-hArl8b. The mixture was inverted, incubated for 10 min at ambient temperature and dropwise added to cells on a 10 cm cell culture dish in 10 mL of DMEM/FBS. After 16 h, the medium was replaced by new DMEM/FBS and cells were incubated for another 24 h before purification of FF compartments after 30 min/120 min (pulse/chase). Six μg of purified compartments were adjusted to 500 μL by addition of HB containing 2 mg/mL BSA and spun onto glass coverslips (1,800 × g, 15 min, 4°C) in a 24-well plate. Supernatants were discarded and compartments were fixed with 4% FA in HB for 16 h at 4°C. FA was discarded and 50 mM NH4Cl in HB was added for 15 min (ambient temperature). Samples were blocked with 4% (w:v) goat serum in PBS (blocking buffer) for 30 min, incubated with a rabbit anti-Rab7 antibody (ab137029,66 diluted 1:50 per volume in blocking buffer) for 60 min, washed five times with PBS and incubated with an Alexa488-labeled secondary antibody (diluted 1:200 per volume in blocking buffer) for 30 min. Coverslips were washed five times in PBS and once in ddH2O and mounted in 2 μL mowiol. Samples were analyzed using a Zeiss Axio Observer epifluorescence microscope equipped with a 100x oil immersion objective and a HXP lamp.
HeLa WT cells in 10 cm dishes were transfected with pEGFP-C1-Rab7a T22N using jetPRIME reagent (Polyplus) according to the manufacturer’s protocol.
Curve fitting
In HeLa cells, all proteins tested accumulated on FF endosomes over time, neither of them was lost within the interval analyzed (Figures 3A–3F). Hence, the amount of each protein on endosomes at 120 min corresponded to the maximum protein amount acquired and was set as 100%. The kinetics of protein acquisition by FF endosomes were best described by the logistic function g(x, a) which hence was used for curve fitting.
For each protein analysed, the constants a1, a2, and a3 within g(x, a) were determined according to the least squares method by vector analysis including a Nabla-operator and the Levenberg-Marquardt algorithm (PTC Mathcad 14).
QUANTIFICATION AND STATISTICAL ANALYSIS
Mean values and standard deviations were calculated from independent experiments using GraphPad Prism 9. The number of independent experiments is given in the figure legends. All error bars represent standard deviations. No data set was excluded.
Supplementary Material
Highlights.
Endosomes recruit Rab7, Rab2, and HOPS before fusion with Arl8-containing lysosomes
HOPS is recruited to late endosomes by Rab2, not by Rab7
Sites of endosome-lysosome tethering/fusion contain HOPS and Rab2 but not Rab7
HOPS bridges Rab2 on late endosomes with Arl8/BORC on lysosomes
ACKNOWLEDGMENTS
We thank G. Jeschke for help with mathematics, S. Held and A. Jünger-Leif for expert technical assistance, and M. Sharma for plasmids. We thank the Deutsche Forschungsgemeinschaft e.V. (DFG grants HA1929/1-1 to A.H. and SA683/10-1 to P.S.) and the Intramural Program of NICHD, NIH (project ZIA-HD001607) for financial support.
Footnotes
DECLARATION OF INTERESTS
The authors declare that there is no conflict of interest.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111969.
REFERENCES
- 1.Bright NA, Davis LJ, and Luzio JP (2016). Endolysosomes are the principal intracellular sites of acid hydrolase activity. Curr. Biol. 26, 2233–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen PJ, and Steinberg F (2018). To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696. [DOI] [PubMed] [Google Scholar]
- 3.Rink J, Ghigo E, Kalaidzidis Y, and Zerial M (2005). Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749. [DOI] [PubMed] [Google Scholar]
- 4.Gruenberg J, and Stenmark H (2004). The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5, 317–323. [DOI] [PubMed] [Google Scholar]
- 5.Huotari J, and Helenius A (2011). Endosome maturation. EMBO J. 30, 3481–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marwaha R, Arya SB, Jagga D, Kaur H, Tuli A, and Sharma M (2017). The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J. Cell Biol. 216, 1051–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khatter D, Raina VB, Dwivedi D, Sindhwani A, Bahl S, and Sharma M (2015). The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J. Cell Sci. 128, 1746–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bucci C, Thomsen P, Nicoziani P, McCarthy J, and van Deurs B (2000). Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherfils J, and Zeghouf M (2013). Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309. [DOI] [PubMed] [Google Scholar]
- 10.Langemeyer L, Borchers A-C, Herrmann E, Fu€llbrunn N, Han Y, Perz A, Auffarth K, Kümmel D, and Ungermann C (2020). A conserved and regulated mechanism drives endosomal Rab transition. Elife 9, e56090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, and Ungermann C (2010). The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659. [DOI] [PubMed] [Google Scholar]
- 12.Dehnen L, Janz M, Verma JK, Psathaki OE, Langemeyer L, Fröhlich F, Heinisch JJ, Meyer H, Ungermann C, and Paululat A (2020). A trimeric metazoan Rab7 GEF complex is crucial for endocytosis and scavenger function. J. Cell Sci. 133, jcs247080. [DOI] [PubMed] [Google Scholar]
- 13.Poteryaev D, Datta S, Ackema K, Zerial M, and Spang A (2010). Identification of the switch in early-to-late endosome transition. Cell 141, 497–508. [DOI] [PubMed] [Google Scholar]
- 14.Pu J, Schindler C, Jia R, Jarnik M, Backlund P, and Bonifacino JS (2015). BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell 33, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niwa S, Tao L, Lu SY, Liew GM, Feng W, Nachury MV, and Shen K (2017). BORC regulates the axonal transport of synaptic vesicle precursors by activating ARL-8. Curr. Biol. 27, 2569–2578.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, and Neefjes J (2007). Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 176, 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosa-Ferreira C, and Munro S (2011). Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell 21, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keren-Kaplan T, and Bonifacino JS (2021). ARL8 relieves SKIP autoinhibition to enable coupling of lysosomes to kinesin-1. Curr. Biol. 31, 540–554.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guardia CM, Fariás GG, Jia R, Pu J, and Bonifacino JS (2016). BORC functions upstream of kinesins 1 and 3 to coordinate regional movement of lysosomes along different microtubule tracks. Cell Rep. 17, 1950–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia R, Guardia CM, Pu J, Chen Y, and Bonifacino JS (2017). BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy 13, 1648–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Kant R, Jonker CTH, Wijdeven RH, Bakker J, Janssen L, Klumperman J, and Neefjes J (2015). Characterization of the mammalian CORVET and HOPS complexes and their modular restructuring for endosome specificity. J. Biol. Chem. 290, 30280–30290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Yang T, Wang S, Wang Z, Yun Y, Sun L, Zhou Y, Xu X, Akazawa C, Hong W, and Wang T (2015). RILP interacts with HOPS complex via VPS41 subunit to regulate endocytic trafficking. Sci. Rep. 5, 8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickner W (2010). Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26, 115–136. [DOI] [PubMed] [Google Scholar]
- 24.Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, Veerapen N, Besra GS, Hacohen N, and Brenner MB (2011). Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity 35, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al. (2015). PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 57, 39–54. [DOI] [PubMed] [Google Scholar]
- 26.Jongsma ML, Bakker J, Cabukusta B, Liv N, van Elsland D, Fermie J, Akkermans JL, Kuijl C, van der Zanden SY, Janssen L, et al. (2020). SKIP-HOPS recruits TBC1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport. EMBO J. 39, e102301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lőrincz P, Tóth S, Benkő P, Lakatos Z, Boda A, Glatz G, Zobel M, Bisi S, Hegedűs K, Takáts S, et al. (2017). Rab2 promotes autophagic and endocytic lysosomal degradation. J. Cell Biol. 216, 1937–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita N, Huang W, Lin T-H, Groulx J-F, Jean S, Nguyen J, Kuchitsu Y, Koyama-Honda I, Mizushima N, Fukuda M, and Kiger AA (2017). Genetic screen in Drosophila muscle identifies autophag-ymediated T-tubule remodeling and a Rab2 role in autophagy. Elife 6, e23367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim BY, Krämer H, Yamamoto A, Kominami E, Kohsaka S, and Akazawa C (2001). Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J. Biol. Chem. 276, 29393–29402. [DOI] [PubMed] [Google Scholar]
- 30.Kim BY, Ueda M, Kominami E, Akagawa K, Kohsaka S, and Akazawa C (2003). Identification of mouse Vps16 and biochemical characterization of mammalian class C Vps complex. Biochem. Biophys. Res. Commun. 311, 577–582. [DOI] [PubMed] [Google Scholar]
- 31.Pirooz SD, He S, Zhang T, Zhang X, Zhao Z, Oh S, O’Connell D, Khalilzadeh P, Amini-Bavil-Olyaee S, Farzan M, and Liang C (2014). UVRAG is required for virus entry through combinatorial interaction with the class C-Vps complex and SNAREs. Proc. Natl. Acad. Sci. USA 111, 2716–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boda A, Lőrincz P, Takáts S, Csizmadia T, Tóth S, Kovács AL, and Juhász G (2019). Drosophila Arl8 is a general positive regulator of lysosomal fusion events. Biochim. Biophys. Acta Mol. Cell Res. 1866, 533–544. [DOI] [PubMed] [Google Scholar]
- 33.Jeschke A, and Haas A (2018). Sequential actions of phosphatidylinositol phosphates regulate phagosome-lysosome fusion. Mol. Biol. Cell 29, 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stroupe C, Collins KM, Fratti RA, and Wickner W (2006). Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 25, 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann I, and Munro S (2006). An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119, 1494–1503. [DOI] [PubMed] [Google Scholar]
- 36.Becken U, Jeschke A, Veltman K, and Haas A (2010). Cell-free fusion of bacteria-containing phagosomes with endocytic compartments. Proc. Natl. Acad. Sci. USA 107, 20726–20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H-S, Stolz DB, and Romero G (2005). Characterization of endocytic vesicles using magnetic microbeads coated with signalling ligands. Traffic (Copenhagen, Denmark) 6, 324–334. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K, Mashima H, Miura K, Maeda D, Goto A, Goto T, Sun-Wada G-H, Wada Y, and Ohnishi H (2017). Disruption of small GTPase Rab7 exacerbates the severity of acute pancreatitis in experimental mouse models. Sci. Rep. 7, 2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saftig P, and Klumperman J (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635. [DOI] [PubMed] [Google Scholar]
- 40.Zhao C, Smith EC, and Whiteheart SW (2012). Requirements for the catalytic cycle of the N-ethylmaleimide-Sensitive Factor (NSF). Biochim. Biophys. Acta 1823, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Höing S, Yeh T-Y, Baumann M, Martinez NE, Habenberger P, Kremer L, Drexler HCA, Küchler P, Reinhardt P, Choidas A, et al. (2018). Dynarrestin, a novel inhibitor of cytoplasmic dynein. Cell Chem. Biol. 25, 357–369.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillingham AK, Sinka R, Torres IL, Lilley KS, and Munro S (2014). Toward a comprehensive map of the effectors of rab GTPases. Dev. Cell 31, 358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kajiho H, Kajiho Y, Frittoli E, Confalonieri S, Bertalot G, Viale G, Di Fiore PP, Oldani A, Garre M, Beznoussenko GV, et al. (2016). RAB2A controls MT1-MMP endocytic and E-cadherin polarized Golgi trafficking to promote invasive breast cancer programs. EMBO Rep. 17, 1061–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chavrier P, Parton RG, Hauri HP, Simons K, and Zerial M (1990). Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62, 317–329. [DOI] [PubMed] [Google Scholar]
- 45.Lund VK, Madsen KL, and Kjaerulff O (2018). Drosophila Rab2 controls endosome-lysosome fusion and LAMP delivery to late endosomes. Autophagy 14, 1520–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, and Klausner RD (1991). Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 67, 601–616. [DOI] [PubMed] [Google Scholar]
- 47.Nakae I, Fujino T, Kobayashi T, Sasaki A, Kikko Y, Fukuyama M, Gengyo-Ando K, Mitani S, Kontani K, and Katada T (2010). The arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol. Biol. Cell 21, 2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Repnik U, Česen MH, and Turk B (2013). The endolysosomal system in cell death and survival. Cold Spring Harbor Perspect. Biol. 5, a008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson DE, Ostrowski P, Jaumouillé V, and Grinstein S (2016). The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasuda S, Morishita S, Fujita A, Nanao T, Wada N, Waguri S, Schiavo G, Fukuda M, and Nakamura T (2016). Mon1-Ccz1 activates Rab7 only on late endosomes and dissociates from the lysosome in mammalian cells. J. Cell Sci. 129, 329–340. [DOI] [PubMed] [Google Scholar]
- 51.van der Welle REN, Jobling R, Burns C, Sanza P, van der Beek JA, Fasano A, Chen L, Zwartkruis FJ, Zwakenberg S, Griffin EF, et al. (2021). Neurodegenerative VPS41 variants inhibit HOPS function and mTORC1-dependent TFEB/TFE3 regulation. EMBO Mol. Med. 13, e13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanlandingham PA, and Ceresa BP (2009). Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284, 12110–12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levin-Konigsberg R, Montaño-Rendón F, Keren-Kaplan T, Li R, Ego B, Mylvaganam S, DiCiccio JE, Trimble WS, Bassik MC, Bonifacino JS, et al. (2019). Phagolysosome resolution requires contacts with the endoplasmic reticulum and phosphatidylinositol-4-phosphate signalling. Nat. Cell Biol. 21, 1234–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heinrich M, Wickel M, Schneider-Brachert W, Sandberg C, Gahr J, Schwandner R, Weber T, Saftig P, Peters C, Brunner J, et al. (1999). Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J. 18, 5252–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pryor PR, Mullock BM, Bright NA, Lindsay MR, Gray SR, Richardson SCW, Stewart A, James DE, Piper RC, and Luzio JP (2004). Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5, 590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis LJ, Bright NA, Edgar JR, Parkinson MDJ, Wartosch L, Mantell J, Peden AA, and Luzio JP (2021). Organelle tethering, pore formation and SNARE compensation in the late endocytic pathway. J. Cell Sci. 134, jcs255463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pols MS, ten Brink C, Gosavi P, Oorschot V, and Klumperman J (2013). The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic (Copenhagen, Denmark) 14, 219–232. [DOI] [PubMed] [Google Scholar]
- 58.Bertram EM, Hawley RG, and Watts TH (2002). Overexpression of rab7 enhances the kinetics of antigen processing and presentation with MHC class II molecules in B cells. Int. Immunol. 14, 309–318. [DOI] [PubMed] [Google Scholar]
- 59.Anderson J, Walker G, and Pu J (2022). BORC-ARL8-HOPS ensemble is required for lysosomal cholesterol egress through NPC2. Mol. Biol. Cell 33, ar81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiani ML, Beitz J, Turvy D, Blum JS, and Stahl PD (1998). Regulation of mannose receptor synthesis and turnover in mouse J774 macrophages. J. Leukoc. Biol. 64, 85–91. [DOI] [PubMed] [Google Scholar]
- 61.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolte S, and Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232. [DOI] [PubMed] [Google Scholar]
- 63.Jeschke A, Zehethofer N, Lindner B, Krupp J, Schwudke D, Haneburger I, Jovic M, Backer JM, Balla T, Hilbi H, and Haas A (2015). Phosphatidylinositol 4-phosphate and phosphatidylinositol 3-phosphate regulate phagolysosome biogenesis. Proc. Natl. Acad. Sci. USA 112, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rai A, Pathak D, Thakur S, Singh S, Dubey AK, and Mallik R (2016). Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell 164, 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geumann U, Barysch SV, Hoopmann P, Jahn R, and Rizzoli SO (2008). SNARE function is not involved in early endosome docking. Mol. Biol. Cell 19, 5327–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jimenez-Orgaz A, Kvainickas A, Nägele H, Denner J, Eimer S, Dengjel J, and Steinberg F (2018). Control of RAB7 activity and localization through the retromer-TBC1D5 complex enables RAB7-dependent mitophagy. EMBO J. 37, 235–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


