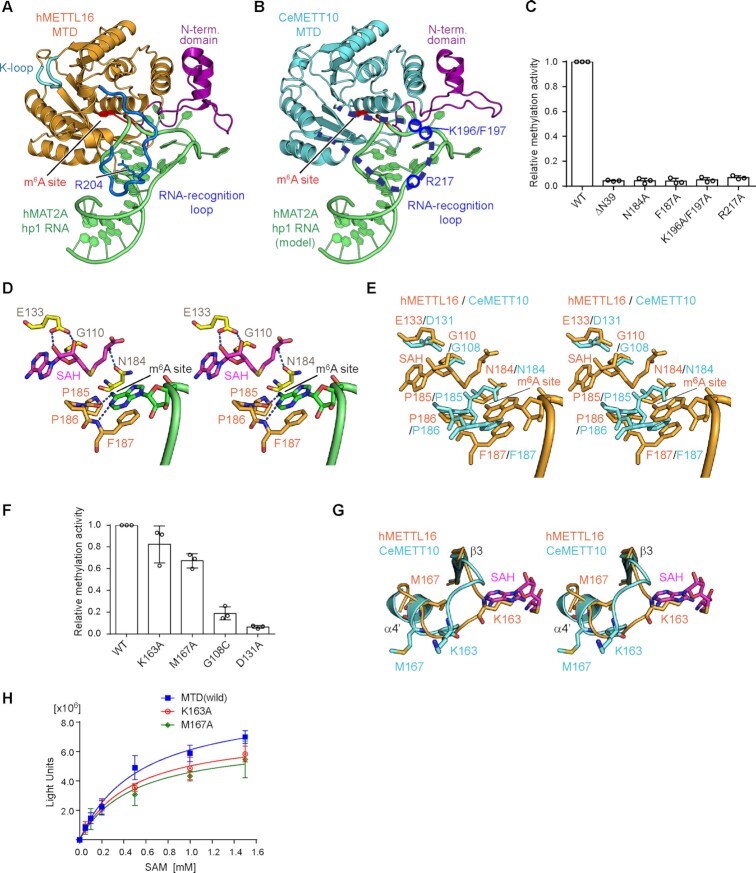
Figure 2.
Key residues of CeMETT10- MTD for RNA methylation. (A) The structure of human METTL16-MTD (hMETTL16-MTD) in complex with hMAT2A- hp1 RNA (PDB: 6DU4) (23). hMAT2A-hp1 is shown in green. (B) RNA docking onto the CeMETT10-MTD structure. The structure of hMETTL16-MTD complexed with hMAT2A-hp1 in (A) was superimposed on the structure of CeMETT10-MTD. hMETTL16-MTD was omitted for clarity. hMAT2A-hp1 is colored green. (C) Methylation of sams-hp RNA (see Figure 3B) by METT10-MTD and its variants. The sams-hp RNA (1 μM) was incubated with 0.4 μM METT10-MTD in the presence of 1 mM SAM, at 37°C for 4 min. The methylation of sams-hp RNA by wild-type METT10-MTD was taken as 1.0. The bars in the graphs are the SD of three independent experiments. (D) Model of the hMETTL16 active site with SAH and hMAT2A-hp1. The SAH was modeled into the structure of hMETTL16-MTD in complex with hMAT2A-hp1 by referring to the structure of hMETTL16-MTD in complex with SAH (PDB: 6B92) (34). The methylation site in the RNA (m6A site) is shown in green, and SAH is colored magenta. (E) Superimposition of the active site structures of CeMETT10 (cyan) and hMETTL16 (orange). (F) Methylation of sams-hp RNA by METT10-MTD and its variants, as in (C). The methylation of sams-hp RNA by wild-type METT10-MTD was taken as 1.0. The bars in the graphs are the SD of three independent experiments. (G) Superimposition of the K-loops of CeMETT10-MTD (cyan) and hMETTL16-MTD (orange) in complex with hMAT2A-hp1. SAH was modeled as in (D). (H) Methylation reactions of sams-hp-ls RNA by METT10-MTD and its variants with various concentrations of SAM (0.05, 0.1, 0.2, 0.5, 1.0 and 1.5 mM). The sams-hp RNA (20 μM) was incubated with 0.4 μM METT10-FLΔL or its variants in the presence of various concentrations of SAM, at 37°C for 4 min. The bars in the graphs are the SD of three independent experiments.