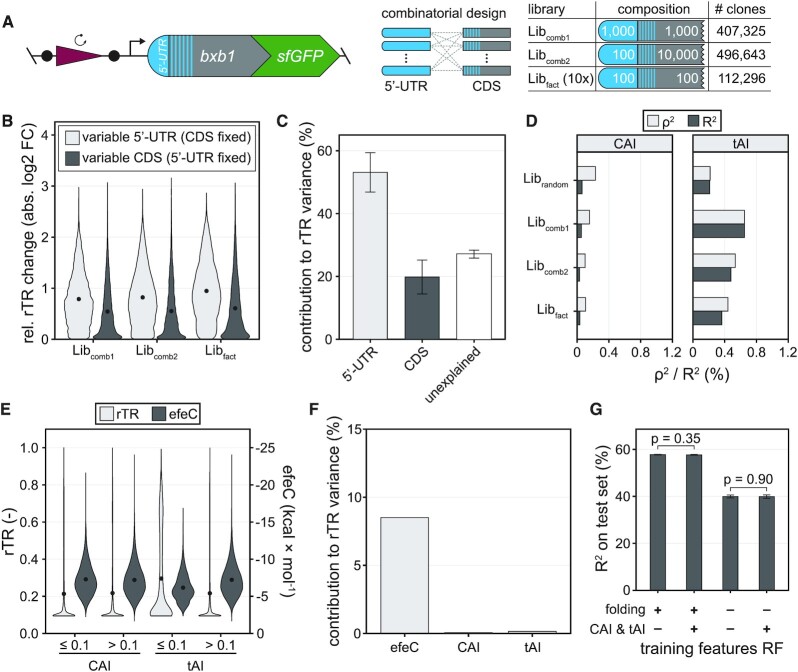
Figure 4.
Overall impact of 5′-UTR, CDS and codon usage on translation initiation. (A) Three additional libraries of combinatorial (Libcomb1, Libcomb2) and full-factorial (Libfact) design were assessed via uASPIre. Libcomb1: combinatorial combination of about 1000 5′-UTRs and 1000 CDSs. Libcomb2: combinatorial combination of about 100 5′-UTRs and 10 000 CDSs. Libfact: ten independent batches, each a full factorial combination of approx. 100 5′-UTRs and 100 CDSs. Libfact was tested in two independent biological replicates. The number of analysed clones is indicated for each library. (B) Impact of the exchange of 5′-UTRs or CDSs on translation initiation. The rTR change (absolute value) of a given 5′-UTR upon exchanging its CDS versus the mean rTR of all variants with that same 5′-UTR is displayed (and vice versa). Black circles within violins are mean relative rTR changes. (C) ANOVA with the mean rTRs of all 5′-UTRs and CDSs in Libfact. Error bars: standard deviation between ten independent batches of Libfact. (D) Correlation of codon usage indices CAI and tAI with rTR. (E) Comparison of rTRs and predicted folding energies (efeC) of variants with low (≤0.1) and high (>0.1) CAI/tAI in all libraries. Black circles within violins are mean rTR/efeC values. (F) Contribution of efeC, CAI and tAI to the rTR variance in all libraries according to an ANOVA with only the three parameters as covariates. (G) Impact of folding and codon usage metrics on the performance of random forest (RF) models trained on Librandom. Sequence parameters for mRNA folding (mfeT, mfeC, efeT, efeC, accT and accC) and codon usage (CAI and tAI) were added or omitted during training. Error bars: Standard deviation of five training repeats with 10-fold cross-validation each. P-values were calculated with Welch two sample t-tests.