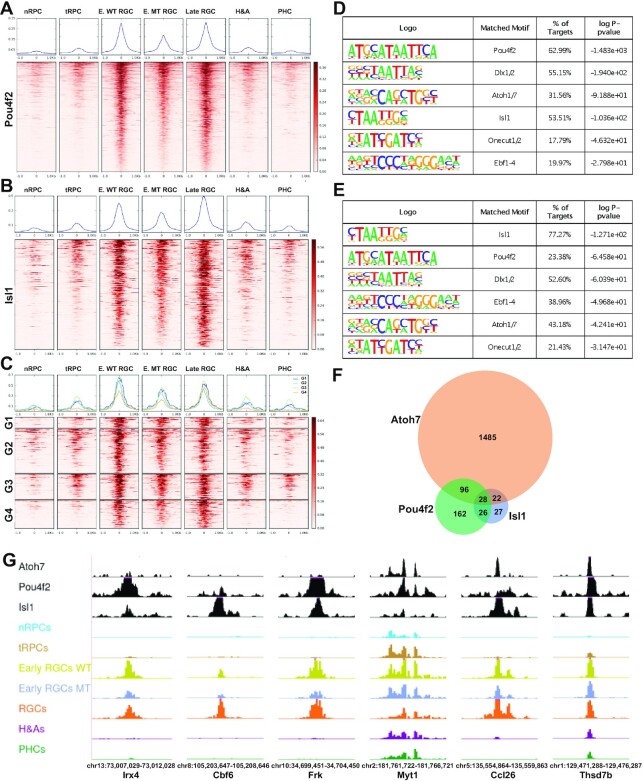
Figure 7.
Pou4f2 and Isl1 bind to RGC-specific enhancers. (A) Activities of Pou4f2-bound enhancers as determined by normalized Tn5 insertion frequencies across different cell states/types at E14.5. (B) Activities of Isl1-bound enhancers across different cell states/types. (C) Activities of enhancers co-bound by at least two of the three transcription factors Atoh7, Pou4f2 and Isl1. G1: Atoh7 + Pou4f2 + Isl1; G2: Atoh7 + Pou4f2; G3: Atoh7 + Isl1; G4: Pou4f2 + Isl1. For (A–C), genomic regions the MACS2 summits ± 1.0 kb were displayed. Color scales represent normalized Tn5 insertion frequencies. Normalized average Tn5 insertion frequencies in the different cell states/types are displayed on top of each heatmap. Pou4f2 summits are used in (C) to generate the heatmap. (D) Motif enrichment analysis by HOMER of Pou4f2-bound enhancers displayed in (A). (E) Motif enrichment analysis by HOMER of Isl1-bound enhancers displayed in (B). (F) A Venn diagram demonstrating the overlaps of genes likely regulated by Atoh7, Pou4f2 and Isl1. (G) Example enhancers bound by Pou4f2 and/or Isl1, with or without Atoh7 binding, and their cell state/type-specific activities at E14.5 as displayed in the genome browser. Note that the activities of all these enhancers were highly RGC specific and significantly reduced in the Atoh7-null early RGCs (Early RGCs MT) as compared to the wild-type early RGCs (early RGCs WT). Genome coordinates (mm10) of the displayed regions and the nearby genes are indicated at the bottom.