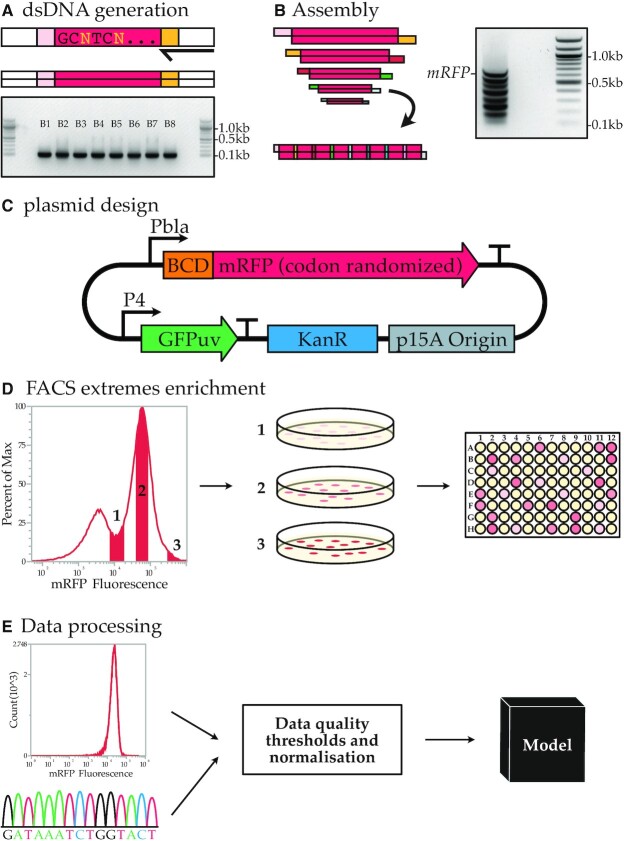
Figure 1.
Codon randomized library generation and analysis. (A) PCR is used to generate dsDNA from oligos and an electrophoresis gel yields the eight dsDNA blocks used to build codon randomized mRFP. (B) The assembly reaction and the electrophoresis gel result of the assembly. The complete assembly of all eight blocks is indicated with the mRFP tag. The seven bands below are intermediate products. (C) The expression vector used to express the codon random mRFP. (D) FACS enrichment for a wide expression range within the library is used to obtain a higher representation of the high and low expressing codon variants. (E) Flow cytometry analysis of cultures and Sanger sequencing data are QA passed and used in machine learning models.