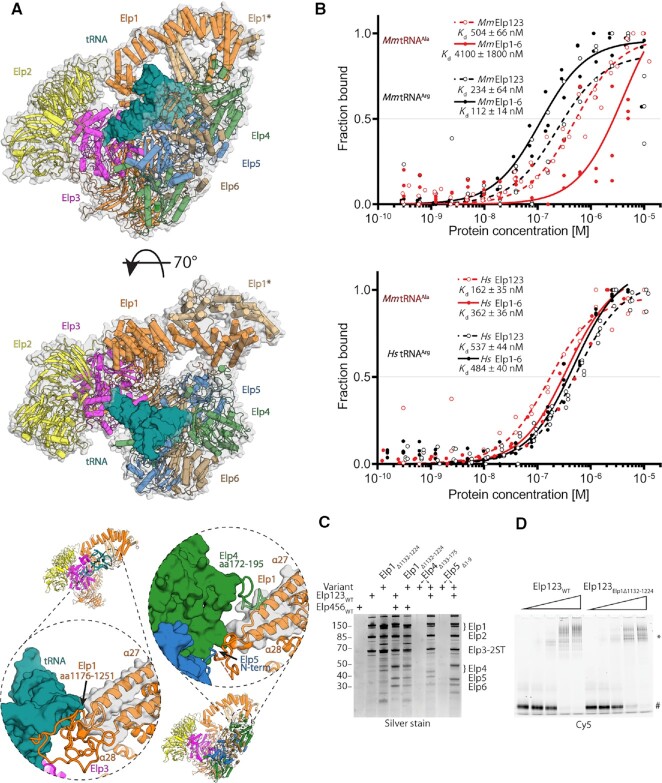
Figure 5.
Biochemical and functional characterization of Elongator complex from higher eukaryotes. (A) Superposition of ScElp123–tRNA and ScElongator in two orientations, showing a possibility of the tRNA (deep teal) binding by the latter complex (above). Below: yeast CP2 comparison between ScElp123–tRNA and ScElongator complexes showing Elp1 tRNA binding loop displacement. (B) Microscale thermophoresis measurements, respective fits, and calculated dissociation constant (Kd) values for the mouse (top) and human (bottom) Elp123 (red) and Elongator (black). In both cases, the Hill coefficient is close to 1, indicating the presence of independent binding sites. n = 3. (C) SDS-PAGE gel comparing in vitro reconstitution of human Elongator complex with the use of Elp1, Elp4 and Elp5 functional variants of CP2 structural features presented in the previous panel. Twin-Strep-tag indicated on Elp3 subunit. (D) EMSA assay using wild type human Elp123 (left), human Elp123Elp1Δ1132–1224 (right), and fluorescently labeled tRNA AlaUGC. The positions of free tRNA and protein–tRNA complexes (*) are indicated next to the native polyacrylamide gel electrophoresis.