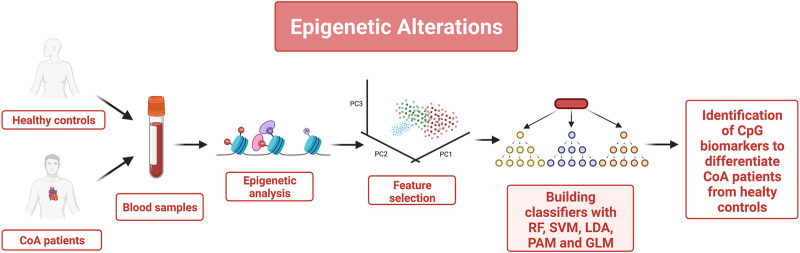
FIGURE 3.
Blood samples were utilized to identify epigenetic changes that allow distinguishing healthy controls from patients with coarctation of the aorta (CoA). Cytosine nucleotide (CpG) methylation changes obtained from Genome-wide DNA methylation analysis were selected as mean features. Only the probes with statistically significant methylation differences combined with a sufficiently high methylation fold change were kept as features and Multivariate approaches such as principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) were used to confirm that these features could accurately distinguish the CoA samples from the controls. 5 classifiers were built and trained using these features. Features with the highest predictive capabilities are potential novel biomarkers candidates for CoA.