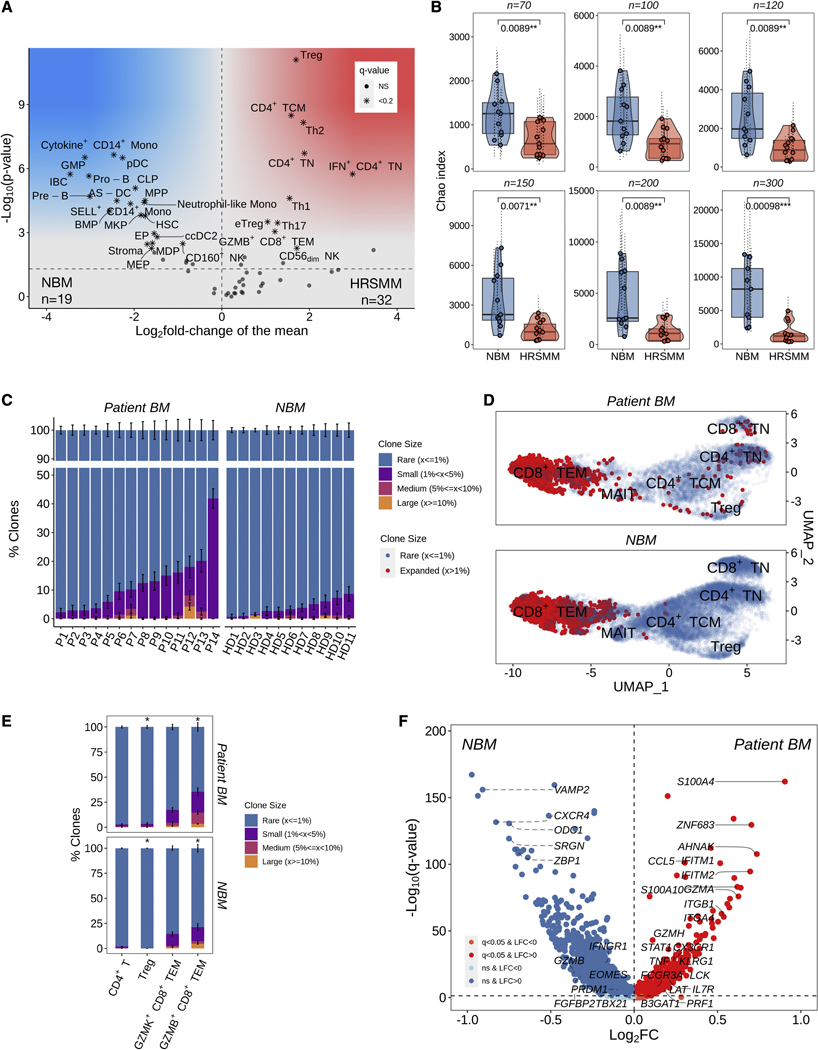
Figure 3.
Comprehensive profiling of changes in BM immune cell composition and TCR repertoire in patients with HRSMM. A) Volcano plot of proportion changes in the BM of patients with HRSMM (n=32) compared to HD (NBM, n=19) with at least 100 cells overall. P-values were computed with Wilcoxon’s rank-sum test and corrected using the Benjamini-Hochberg approach. Cell types with a q-value of <0.2 were marked with stars. B) Boxplots, violin plots, and scatter plots comparing BM TCR repertoire diversity, as assessed by the Chao index, between patients with HRSMM (n=14) and healthy individuals (NBM, n=11) given different numbers of downsampled cells. Each data point represents the average diversity estimate across 100 random samples of the given size for a single sample; the range of diversity estimates across all iterations for each sample is visualized in error bars (dotted line). Violin outline width represents density. P-values were computed with Wilcoxon’s rank-sum tests. (Box: 1st quartile, median, 3rd quartile; whiskers: +/− 1.5*IQR). C) Barplots showing the proportion of T cells in a given BL patient sample (P, n=14) or sample from a HD (n=11) that were determined to belong to one of four clone size categories (Rare: ≤1%; Small: >1% and <5%; Medium: ≥5% and <10%; Large: ≥10%) through iterative (n=100) downsampling of 100 cells. The average proportion per clone size category was visualized and the standard deviation across iterations was depicted in solid-line error bars. D) UMAP embedding of HD BM (NBM) and patient BM T cells at BL with matched TCR data. T cells belonging to rare clonotypes (with a frequency of ≤1%) were colored in blue, while T cells belonging to expanded clonotypes (with a frequency of >1%) were colored in red. E) Barplots showing the proportion of clonotypes in a given T cell subtype across all patients (n=14) or HD (n=11) that belonged to one of the four clone size categories. For each T cell subtype, 100 cells were randomly sampled 100 times from all patients or HD, and the proportion of expanded (1-Rare) clonotypes was compared between patients and HD using bootstrapping with 10,000 iterations. The average proportion per clone size category was visualized and the standard deviation across iterations was depicted in solid-line error bars. F) Volcano plot highlighting genes that are highly expressed in expanded GZMB+ CD8+ TEM cells from patients (n=743, in red) compared to expanded GZMB+ CD8+ TEM cells from HD (n=1,074, in blue). See also Figures S3 and S4.