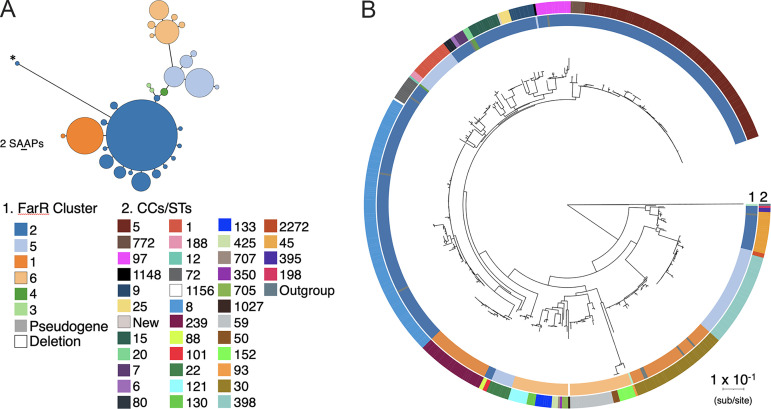
FIG 1.
Grapes plot of six major FarR clusters and associated variants (A) and mapping of FarR clusters on a proteome based Staphylococcus aureus phylogenetic map (B). For the Grapes plot (A), the minimum spanning tree was constructed by alignment of 564 full-length FarR proteins using GrapeTree, which identified 6 primary clusters and associated variants. Points represent groups of identical elements, with point size correlated with number of elements on a log scale. Phylogenetic distance is scaled to two single-amino acid polymorphisms (2SAAP). The asterisk on cluster 2 (*) marks FarR variant 2c where duplication of an amino acid codon at E93 alters the alignment. For the phylogenetic map (B), conceptually translated nucleotide sequences available from PATRIC are shown for 574 S. aureus and 1 S. argenteus strains using PhyloPhlAn3. Initial phylogenetic analysis used FastTree, which was refined with RaxML. The six primary FarR clusters are mapped on ring 1, while clonal complex associations are mapped on ring 2. The colored legend for clonal complex designations is presented in the same order of appearance as on ring 2, beginning with CC5 and progressing in descending order in each column from left to right, ending with the S. argenteus outgroup.