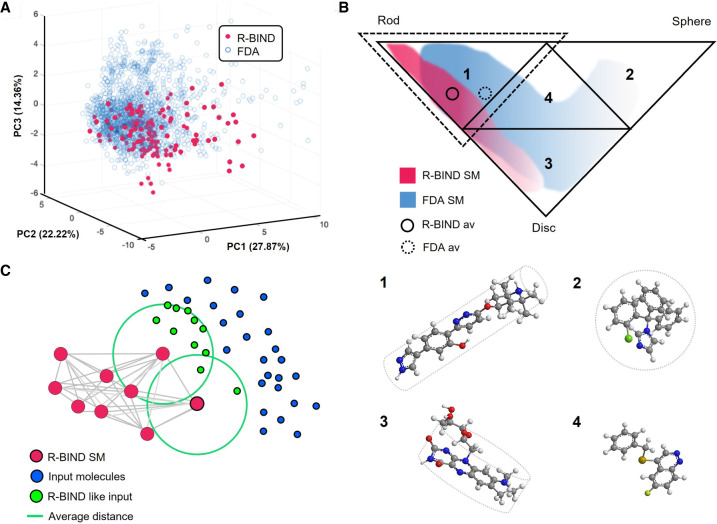
FIGURE 2.
(A) 3D representation of principal components (PCs) 1, 2, and 3 that plots R-BIND SMs and FDA-approved SMs (adapted with permission from Donlic et al. 2022, © American Chemical Society; (B) Principal moments of inertia (PMI) triangle partitions in four subtriangles, representing rod-like (1), sphere-like (2), disc-like (3) and hybrid shapes (4); example molecules are provided for each shape, (1) NVS-SM1 (Palacino et al. 2015), (2) compound 139 from FDA curated library (Donlic et al. 2022), (3) roseoflavin (Lee et al. 2009), and (4) CP6 (Khan et al. 2019) (adapted with permission from Morgan et al. 2017, © Wiley VCH); the dashed triangle outlines the subtriangle 1 where library averages are located. (C) Schematic representation of the k-nearest neighbor (k-NN) algorithm. The R-BIND SMs, plotted in the chemical space, are used to define nearest neighbors, averaging the smallest distance for each new molecule (adapted with permission from Morgan et al. 2019, © American Chemical Society).