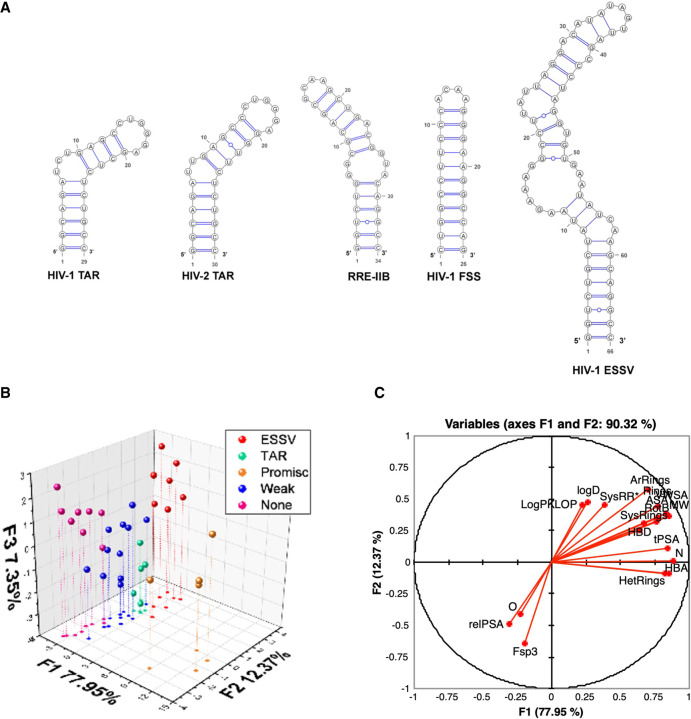
FIGURE 5.
(A) Secondary structures of HIV RNAs screened with DMA library. (TAR) Trans-activation response element, (RRE) rev response element, (FSS) frameshift-stimulating, (ESSV) exonic splicing silencer of Vpr. (B) Linear discriminate analysis (LDA) plot based on 20 cheminformatic parameters clusters to differentiate five groups of ligands for HIV RNA targets. (C) LDA loading plot for the qualitative analysis of the contribution of each cheminformatic parameter contributing on F1 versus F2. (MW) Molecular weight, (HBA) number of hydrogen bond acceptors, (HBD) number of hydrogen bond donors, (LogP) n-octanol/water partition coefficient, (RotB) number of rotatable bonds, (tPSA) topological polar surface area, (LogD) n-octanol/water distribution coefficient, (N) number of nitrogen atoms, (O) number of oxygen atoms, (Rings) number of rings, (ArRings) number of aromatic rings, (HetRings) number of heteroatom-containing rings, (SysRings) number of ring systems, (SysRR) ring complexity, (Fsp3) fraction of sp3 hybridized carbons, (ASA) accessible surface area, (relPSA) relative polar surface area, (VWSA) van der Waals surface area (panels A–C adapted with permission from Patwardhan et al. 2019b, © Royal Society of Chemistry).