FIGURE 1.
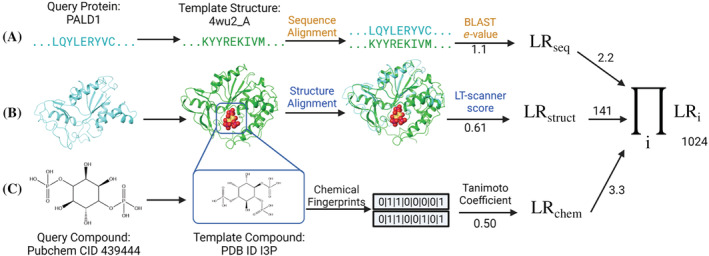
PrePCI algorithm for the prediction of protein‐compound interactions. PrePCI uses BLAST and LT‐scanner for the query protein (aqua sequence and model, left) to identify proteins within PDB protein‐compound complexes that have (a) sequence and/or (b) structural similarity to the query. (c) Compounds predicted to bind the query are identified using a Tanimoto coefficient (TC) chemical similarity search of fingerprints representing the PDB compound and compounds from PubChem. In this example, the sequence and model for Paladin (PALD1) are matched to the PDB complex (PDB ID 4wu2) of Selenomonas ruminatum myo‐inositol hexaphosphate phosphohydrolase bound to the PDB compound I3P with BLAST e‐value 1.1 and LT‐scanner score 0.61. A query compound from PubChem (CID 439444) has TC = 0.5 with the PDB compound. The LR for the interaction between PALD1 and the query compound is the product of the LRs from sequence, structure, and chemical similarity scores.
