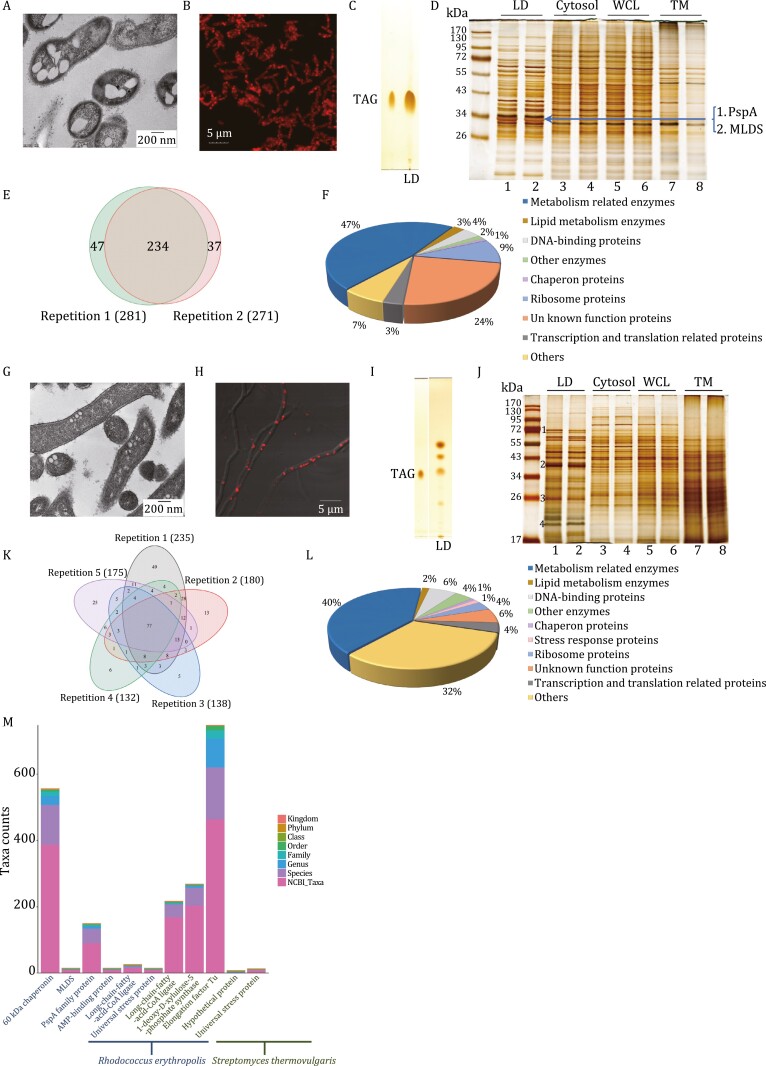
Figure 2.
Identification and analysis of LDs in cloned gut bacteria. (A–D) A strain of human gut bacteria, Rhodococcus erythropolis, was used for analysis of LD in gut bacteria. R. erythropolis was cultured in MSM medium. (A) Ultra-thin structural analysis of R. erythropolis by TEM. Briefly, the bacteria were fixed immediately after collection, dehydrated, and embedded, and finally sectioned into ultra-thin sections (70 nm). The sections were stained and viewed by TEM. Scale bar, 200 nm. (B) Confocal images of R. erythropolis were captured after neutral lipid staining with LipidTOX Red. Scale bar, 5 µm. LDs were then isolated from R. erythropolis using the method we previously reported (Ding et al., 2012). (C) Neutral lipids of isolated LDs were analyzed by thin layer chromatography (TLC). Lane 1, TAG marker; Lane 2, LDs of R. erythropolis. Then the proteins of the LDs isolated from R. erythropolis were studied through biochemical methods and proteomic analysis. (D) The protein profiles of the cellular fractions of R. erythropolis were analyzed by SDS–PAGE and silver staining. The major band in the gel indicated was sliced and subjected to in-gel digestion followed by LC/MS/MS identification. TM, total membrane; WCL, whole cell lysate. (E and F) Two LD isolations from two independent cultures of R. erythropolis were conducted. The total LD proteins of each isolation were subjected to proteomic identification. (E) A Venn diagram was built to present the overlap of the identified LD proteins of the two biological replicates. (F) The identified LD proteins from LC/MS/MS were then analyzed using bioinformatics tools and the proteins with two unique peptides or above were selected. Based on their functions, they were categorized into nine groups. (G–J) The other strain of human gut bacteria, Streptomyces thermovulgaris, was used for analysis of LD in gut bacteria. S. thermovulgaris was cultured in ISP2 medium. (G) Ultra-thin structural analysis of S. thermovulgaris by TEM. Briefly, the bacteria were fixed immediately after collection, dehydrated, embedded, and finally sectioned into ultra-thin sections (70 nm). The sections were stained and viewed by TEM. Scale bar, 200 nm. (H) Confocal images of S. thermovulgaris were captured after neutral lipid staining with LipidTOX Red. Scale bar, 5 µm. LDs were isolated. (I) Neutral lipids of isolated LDs were analyzed by TLC. Lane 1, TAG marker; Lane 2, LDs of S. thermovulgaris. The proteins of the isolated LDs from S. thermovulgaris were studied using biochemical methods and proteomic analysis. (J) The protein profiles of the cellular fractions of S. thermovulgaris were analyzed by SDS–PAGE and silver staining. The four major bands indicated were sliced and subjected to in-gel digestion followed by LC/MS/MS identification. TM, total membrane; WCL, whole cell lysate. (K and L) Five LD isolations from five independent cultures of S. thermovulgaris were conducted. The total LD proteins of each isolation were subjected to proteomic identification independently. (K) A Venn diagram was built to present the overlap of the identified LD proteins of the five biological replicates. (L) The identified LD proteins from LC/MS/MS were then analyzed using bioinformatics tools and the proteins with two unique peptides or above were selected. Based on their functions, they were categorized into 10 groups. (M) Taxonomic dissection and informatics analysis of several LD abundant proteins of gut bacteria. The taxonomic composition distribution histograms at each classification level of the LD abundant proteins from gut bacteria R. erythropolis and S. thermovulgaris are presented. The Kraken2 database was used and the sequences with identity values above or equal to 80% were screened.