FIGURE 1.
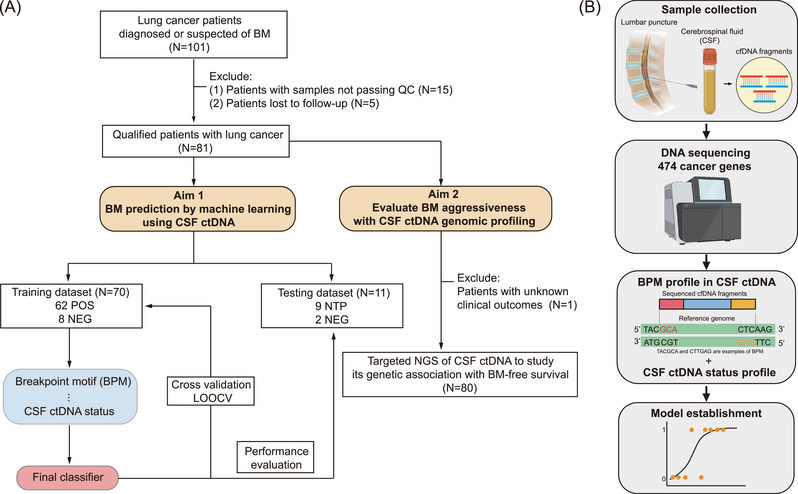
Schematic demonstration of the study design. (A) After excluding 15 patients with CSF samples that failed to pass the quality control (QC) examination and five patients lost to follow‐up, sequencing data of 81 patients were analyzed to assess the potential of cerebrospinal fluid (CSF) circulating tumour DNA (ctDNA) in brain metastases (BM) prediction. Based on the BM status, these 81 patients were categorized into three subgroups: NEG, NTP and POS. The BM predictive model was developed based solely on the breakpoint motif (BPM) profile (BPM model) or combined CSF ctDNA status and BPM profile (integrated model). To train the machine learning model, 62 POS and eight randomly selected NEG patients were recruited into the training dataset. Leave‐one‐out cross‐validation (LOOCV) was performed to evaluate the predictive performance of the BM predictive model, and the area under the curve (AUC) was calculated based on the holdout predictions. Independent testing of the model's performance was performed in the testing cohort comprising nine NTP and two randomly selected NEG patients. A higher predicted score indicates a higher probability of developing BM. Meanwhile, targeted next‐generation sequencing (NGS) data of CSF ctDNA in 80 patients with known clinical outcome data were assessed to reveal genetic alterations related to BM aggressiveness. (B) Schematic demonstration of the integrated model construction. CSF samples were obtained from each participant through the lumbar puncture. A total of 474 cancer‐related genes in CSF ctDNA were sequenced using high‐throughput targeted NGS. The BPM profile was combined with each patient's CSF ctDNA status to generate the integrated machine‐learning model using the elastic‐net logistic regression algorithm.
