FIGURE 4.
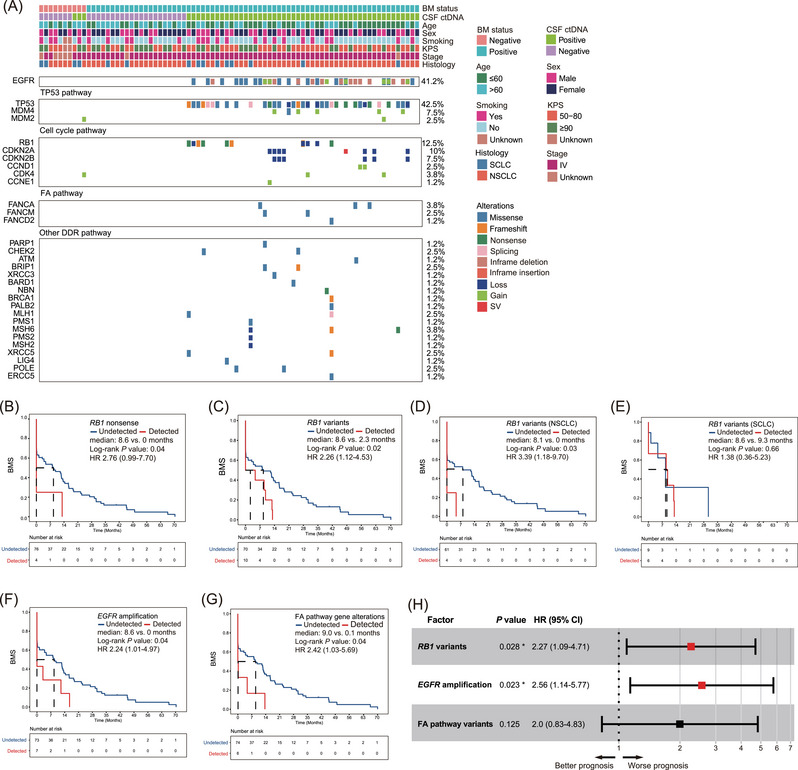
Identifying brain metastases (BM)‐associated genetic alterations using cerebrospinal fluid (CSF) circulating tumour DNA (ctDNA). (A) Co‐mutation plot of lung cancer patients with confirmed or suspected BM (N = 80). Patients’ clinical characteristics are shown on the top, while the top frequently mutated gene alterations are grouped by oncogenic pathways previously reported to be associated with LCBM. (B) Kaplan Meier estimates of BM‐free survival (BMS) in lung cancer patients harbouring RB1 nonsense mutations compared to those without detectable RB1 nonsense mutations. (C) BMS in lung cancer patients harbouring RB1 variants (including SNVs and CNVs) compared to patients without detectable RB1 variants. (D and E) BMS in two stratified patient cohorts comprising either NSCLC (D) or SCLC patients (E) with co‐occurring RB1 variants compared to corresponding control patients. (F) BMS in lung cancer patients harbouring EGFR copy‐number gain variants compared to patients without detectable EGFR amplification. (G) BMS in lung cancer patients harbouring Fanconi anaemia (FA) pathway genetic alterations compared to others. (H) Multivariate analysis of genetic factors associated with BM. FA pathway variants include missense mutations in FANCA, FANCM and FANCD2. Asterisks represent significant P‐values based on the Cox regression model.
