Abstract
The degradation of macromolecules and organelles by the process of autophagy is critical for cellular homeostasis and is often compromised during aging and disease. Beclin1 and Beclin2 are implicated in autophagy induction, and these homologues share a high degree of amino acid sequence similarity but have divergent N-terminal regions. Here, we investigated the functions of the Beclin homologues in regulating autophagy and mitophagy, a specialized form of autophagy that targets mitochondria. Both Beclin homologues contributed to autophagosome formation, but a mechanism of autophagosome formation independent of either Beclin homologue occurred in response to starvation or mitochondrial damage. Mitophagy was compromised only in Beclin1-deficient HeLa cells and mouse embryonic fibroblasts due to defective autophagosomal engulfment of mitochondria, and the function of Beclin1 in mitophagy required the phosphorylation of the conserved Ser15 residue by the kinase Ulk1. Mitochondria-ER associated membranes (MAMs) are important sites of autophagosome formation during mitophagy, and Beclin1, but not Beclin2 or a Beclin1 mutant that could not be phosphorylated at Ser15, localized to MAMs during mitophagy. Our findings establish a regulatory role for Beclin1 in selective mitophagy by initiating autophagosome formation adjacent to mitochondria, a function facilitated by Ulk1-mediated phosphorylation of Ser15 in its distinct N-terminal region.
INTRODUCTION
Autophagy is a tightly regulated intracellular degradation pathway that is responsible for maintaining energy levels during starvation by providing cells with recycled nutrients. Autophagy is also an important cellular quality control mechanism involved in removing protein aggregates and damaged organelles (1). Enhancing autophagy can protect against disease and extend lifespan (2). In contrast, defects in autophagy are associated with development of different diseases including cancers, neurodegeneration and heart failure (2). Thus, because of its promising therapeutic potential, there is strong interest in delineating the mechanisms involved in regulating autophagy.
Autophagosomes are produced on demand and start with the formation of a phagophore membrane that grows into a mature membrane that engulfs cytoplasmic cargo. The double-membrane autophagosome that forms after cargo sequestration fuses with a lysosome where the content is degraded and recycled (2). Beclin1 regulates autophagy and is involved in initiating the formation of autophagosomes in cells. It is a component of the class III PI3K complex that is responsible for generating phosphatidylinositol 3-phosphate (PI3P) at the site of membrane formation (3, 4). This membrane lipid recruits downstream autophagy effector proteins (5). The site of autophagosome formation is critical in selective autophagy and must occur near the cargo to ensure its efficient engulfment. Despite some progress in understanding this process, the molecular mechanism by which membrane biogenesis is initiated in proximity to the cargo remains elusive.
Autophagy is the main pathway responsible for eliminating damaged or unwanted mitochondria (known as mitophagy). In this process, dysfunctional mitochondria are marked for degradation by the E3 ubiquitin ligase Parkin and then engulfed by autophagosomes for delivery to lysosomes. The specific function of Beclin1 in mitophagy is still uncertain due to conflicting data. One study found that Beclin1 initiates mitophagy by facilitating the translocation of Parkin from cytosol to mitochondria (6). In contrast, another investigation observed that silencing Beclin1 leads to reduced mitophagy without affecting mitochondrial Parkin recruitment (7). A third study reported that mitophagy is independent of Beclin1 (8). Additional studies are needed to clarify the role of Beclin1 in mitophagy. It is also unknown if Beclin2, a mammalian-specific homologue of Beclin1 with ~57% shared sequence identity (9), functions in the mitophagy pathway.
Beclin2 promotes endosomal-mediated degradation of GASP1-dependent GPCRs (9). It is also an inhibitor of innate immune signaling pathways to suppress the production of proinflammatory cytokines (10). Similar to Beclin1, Beclin2 has been implicated in regulating autophagy and is reported to interact with components in the class III PI3K complex (9). Studies in knockout mice further suggest that Beclin1 and Beclin2 have distinct functions. Mice lacking Becn1 die early during embryogenesis (11), whereas Becn2−/− mice are viable but are born at a reduced Mendelian ratio (9). Surviving Becn2−/− mice suffer from splenomegaly, lymphadenopathy, and enhanced inflammatory responses (10). The different phenotypes clearly demonstrate that these two proteins have divergent and likely tissue-specific functions. To date, few studies have focused on Beclin2 and its functions are still relatively unexplored.
In this study, we investigated the functions of Beclin1 and Beclin2 in regulating autophagosome formation and mitophagy. Our findings demonstrated that although Beclin1 and Beclin2 were involved in autophagosome formation under basal conditions, a Beclin1/2-independent mechanism of autophagy was activated in response to starvation or mitochondrial depolarization with FCCP. Also, Beclin1, but not Beclin2, functioned in mitophagy by initiating autophagosome formation at damaged mitochondria. Specifically, our findings suggest that Ulk1-mediated phosphorylation of Ser15 in Beclin1 during mitophagy facilitated its recruitment to mitochondria-ER associated membranes (MAMs) where autophagosome formation is initiated.
RESULTS
Stress-induced autophagosome formation is intact in Beclin1-deficient MEFs
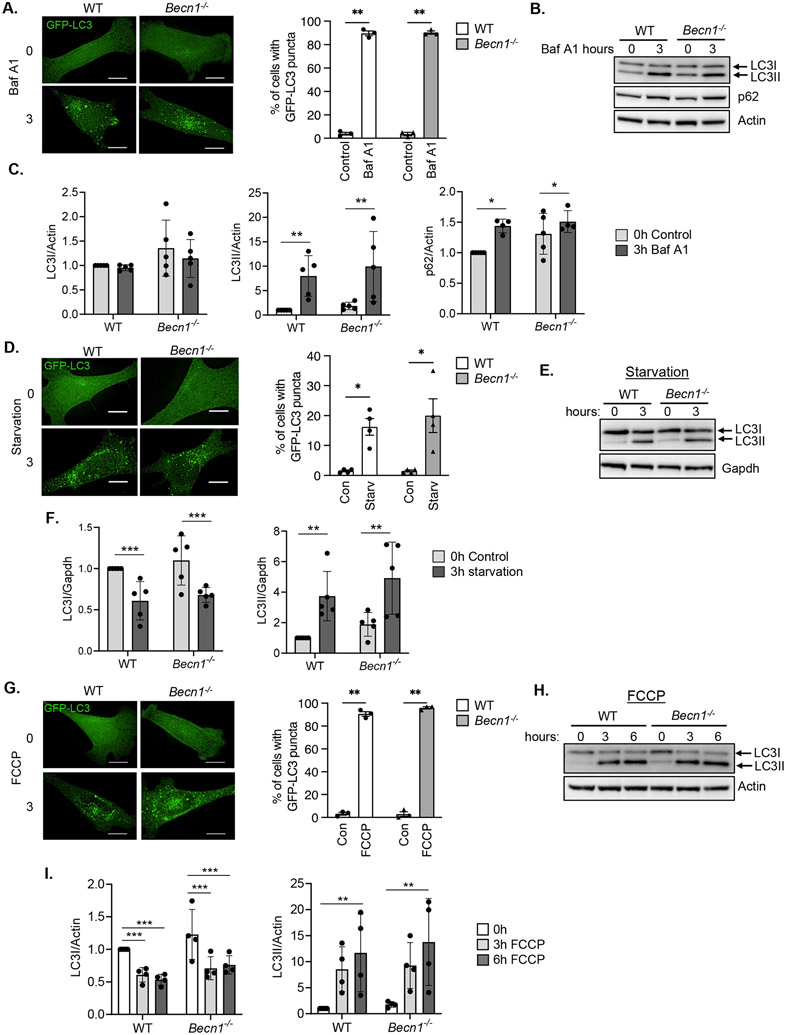
Beclin1 is a component of the class III PI3K complex that initiates the formation of autophagosomes (12). To investigate if autophagosome formation was strictly dependent on Beclin1, we assessed autophagy in wild type (WT) and Becn1−/− mouse embryonic fibroblasts (MEFs). Using GFP-LC3 to visualize autophagosomes, we evaluated basal autophagosome formation in the absence or presence of the v-ATPase inhibitor bafilomycin A1 (Baf A1), which inhibits lysosomal degradation of autophagosomes. The presence of Baf A1 led to accumulation of GFP-LC3-positive vesicles in both WT and Becn1−/− MEFs (Fig. 1A), suggesting that Beclin1 was not essential for the formation of autophagosomes under basal conditions. As an additional measure of autophagosome formation, we analyzed the conversion of LC3I to LC3II by Western blot analysis. Lipidated LC3II is incorporated into the membrane and the amount of LC3II correlates with the number of autophagosomes (13). We also examined the turnover of p62, an adaptor protein that is also an autophagy substrate (13). Western blotting confirmed that treatment with Baf A1 led to increased accumulation of LC3II and p62 in both genotypes (Fig. 1B-C, fig. S12). The levels of cytosolic LC3I were similar in WT and Becn1−/− MEFs basally and in the presence of Baf A1 (Fig. 1B-C, fig. S12). Overall, these findings indicate intact autophagic flux in Becn1−/− MEFs at baseline.
Fig. 1. Formation of autophagosomes at baseline and in response to stress in Becn1−/− deficient MEFs.
A) Representative immunofluorescent images and quantification of the total percentage of cells with GFP-LC3 positive vesicles in WT and Becn1−/− MEFs before (0h) or after (3h) bafilomycin A1 (Baf A1) treatment (100nm) (n=100 cells/group from 3 experiments). B) Representative Western blots for LC3 and p62 in WT and Becn1−/− MEFs at baseline (0h) and after (3h) bafilomycin A1 (Baf A1) treatment (100 nM). C) Quantification of LC3 and p62 protein levels. Actin was used as a loading control (n=4-5 independent experiments). D) Representative immunofluorescent images and quantification of the total percentage of cells with GFP-LC3 positive vesicles in WT and Becn1−/− MEFs before (0h) or after (3h) starvation (Starv) (n=100 cells/group from 3 experiments). E) Representative Western blot for LC3 in WT and Becn1−/− MEFs after starvation. F) Quantification of LC3I and LC3II protein levels (n=5 independent experiments). G) Representative immunofluorescent images and quantification of the total percentage of cells with GFP-LC3 positive vesicles in WT and Becn1−/− MEFs before (0h) or after (3h) FCCP treatment (10 μM) (n=100 cells/group from 3 experiments). H) Representative Western blot for LC3 in WT and Becn1−/− MEFs after 10 μM FCCP treatment. I) Quantification of LC3I and LC3II protein levels (n=4 independent experiments). Data are presented as means ± SEM. *p<0.05, **p<0.01, ***p<0.001, and ns = not significant by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Scale bars = 10 μm.
Next, we investigated whether autophagy induction in response to challenge was intact in Becn1−/− cells. We found that serum and glucose starvation increased the formation of GFP-LC3-positive autophagosomes in both WT and Becn1−/− MEFs (Fig. 1D). Western blot analysis confirmed a similar reduction in LC3I levels with a corresponding increase in LC3II levels in WT and Becn1−/− MEFs after starvation (Fig. 1E-F, fig. S12). We also treated cells with the mitochondrial uncoupler trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP) to induce mitophagy (14). Similar to starvation, FCCP treatment led to increased formation of autophagosomes in WT and Becn1−/− MEFs as evaluated by formation of GFP-LC3 positive vesicles and by Western blotting for LC3I/II (Fig. 1G-I, fig. S12). These results demonstrated that autophagosome generation in response to stress was also independent of Beclin1 in MEFs.
Autophagosome formation is intact in BECN1−/− and BECN2−/− HeLa cells
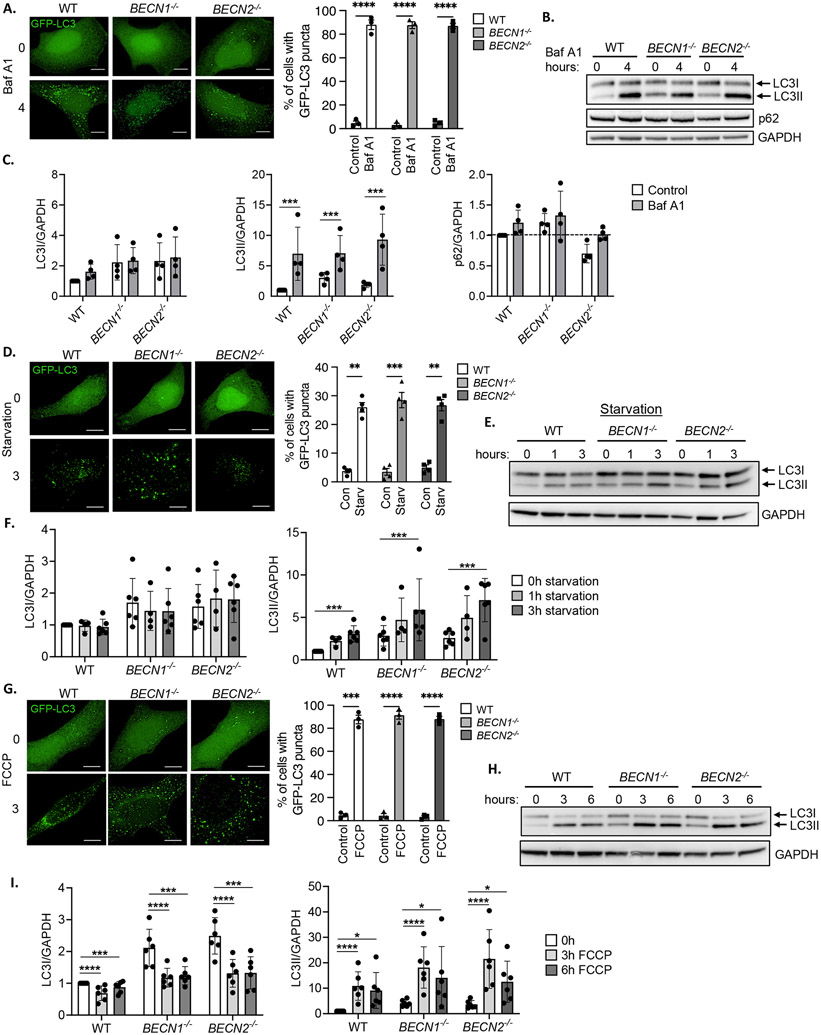
Beclin2, a homolog of Beclin1, has also been implicated in autophagy (9). To further explore the function of Beclin2 in autophagy, we generated BECN1−/− and BECN2−/− HeLa cells by CRISPR-Cas9 gene editing (fig. S1A-C). Consistent with what we observed in MEFs, we found that treatment with Baf A1 led to increased formation of GFP-LC3 positive vesicles in WT, BECN1−/− and BECN2−/− HeLa cells (Fig. 2A), confirming intact autophagosome formation. Western blotting also showed that the accumulation of LC3II was similar in WT, BECN1−/− and BECN2−/− HeLa cells (Fig. 2B-C, fig. S12), indicating a similar rate of autophagosome formation at baseline in the three cell lines. LC3I and p62 levels did not change significantly within this timeframe.
Fig. 2. Formation of autophagosomes in BECN1−/− and BECN2−/− HeLa cells at baseline and in response to stress.
A) Representative immunofluorescent images and quantification of the total percentage of cells with GFP-LC3 positive vesicles in WT, BECN1−/− and BECN2−/− HeLa cells before (0h) or after (4h, 100 nM) bafilomycin A1 (Baf A1) treatment (n=100 cells/group from 3 experiments). B) Representative Western blots for LC3 and p62 in WT, BECN1−/− and BECN2−/− HeLa cells at baseline (0h) and after (4h, 100nM) bafilomycin A1 (Baf A1) treatment. C) Quantification of LC3 and p62 protein levels. GAPDH was used as a loading control (n=4 independent experiments). D) Representative immunofluorescent images and quantification of the total percentage of cells with GFP-LC3 positive vesicles in WT, BECN1−/− and BECN2−/− HeLa cells before or after 3h of starvation (Starv) (n=100 cells/group from 4 experiments). E) Representative Western blots for LC3 in WT, BECN1−/− and BECN2−/− HeLa cells at baseline and after starvation for 1h or 3h. F) Quantification of LC3I and LC3II protein levels (n=4-6 independent experiments). G) Representative immunofluorescent images and quantification of the total percentage of cells with GFP-LC3 positive vesicles in WT, BECN1−/− and BECN2−/− HeLa cells before or after FCCP treatment (10 μM, 3h) (n=100 cells/group from 3 experiments). H) Representative Western blot for LC3 in WT, BECN1−/− and BECN2−/− HeLa cells after 10 μM FCCP treatment for 0, 3, and 6h. I) Quantification of LC3I and LC3II protein levels (n=6 independent experiments). Data are presented as means ± SEM. **p<0.01, ***p<0.001, ****p<0.0001 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Scale bars = 10 μm.
Next, we evaluated whether autophagosome formation in response to challenge was altered in HeLa cells. Nutrient deprivation led to increased formation of GFP-LC3 positive vesicles in WT, BECN1−/− and BECN2−/− HeLa cells as assessed by immunofluorescence analysis (Fig. 2D). Western blotting for LC3II confirmed increased autophagosome formation in all three cell lines in response to starvation (Fig. 2E-F). Moreover, LC3II accumulated in the presence of Baf A1, evidence of intact autophagic flux during starvation (fig. S2A-B). FCCP treatment also led to an increase in GFP-LC3 positive vesicles in WT, BECN1−/− and BECN2−/− HeLa cells (Fig. 2G). Western blotting for LC3I and LC3II confirmed similar FCCP-stimulated autophagosome formation in all three cell lines as assessed by LC3II levels (Fig. 2H-I, fig. S12). Autophagic flux was also intact after extended FCCP exposure (12-24h) in WT, BECN1−/−, and BECN2−/− HeLa cells (fig. S2C-D). The Beclin1-Vps34 and Beclin2-Vps34 complexes are responsible for generating PI3P. To monitor activity of the Vps34 PI3K complexes, we overexpressed a GFP-conjugated PX domain that specifically binds to PI3P (15) in WT, BECN1−/−, and BECN2−/− HeLa cells. We found that FCCP exposure led to a significant increase in localized PI3P generation in all three cell lines (fig. S3A-B). However, the increase in GFP-p40PX punctae in BECN1−/−HeLa cells after FCCP treatment was significantly reduced compared to WT and BECN2−/− HeLa cells, suggesting decreased PI3K activity in Beclin1-deficient cells. Overall, these findings demonstrate that autophagosome formation at baseline and in response to stress occurred independently of Beclin1 and Beclin2 in HeLa cells.
Autophagosome formation in response to stress does not depend on Beclin1 and Beclin2
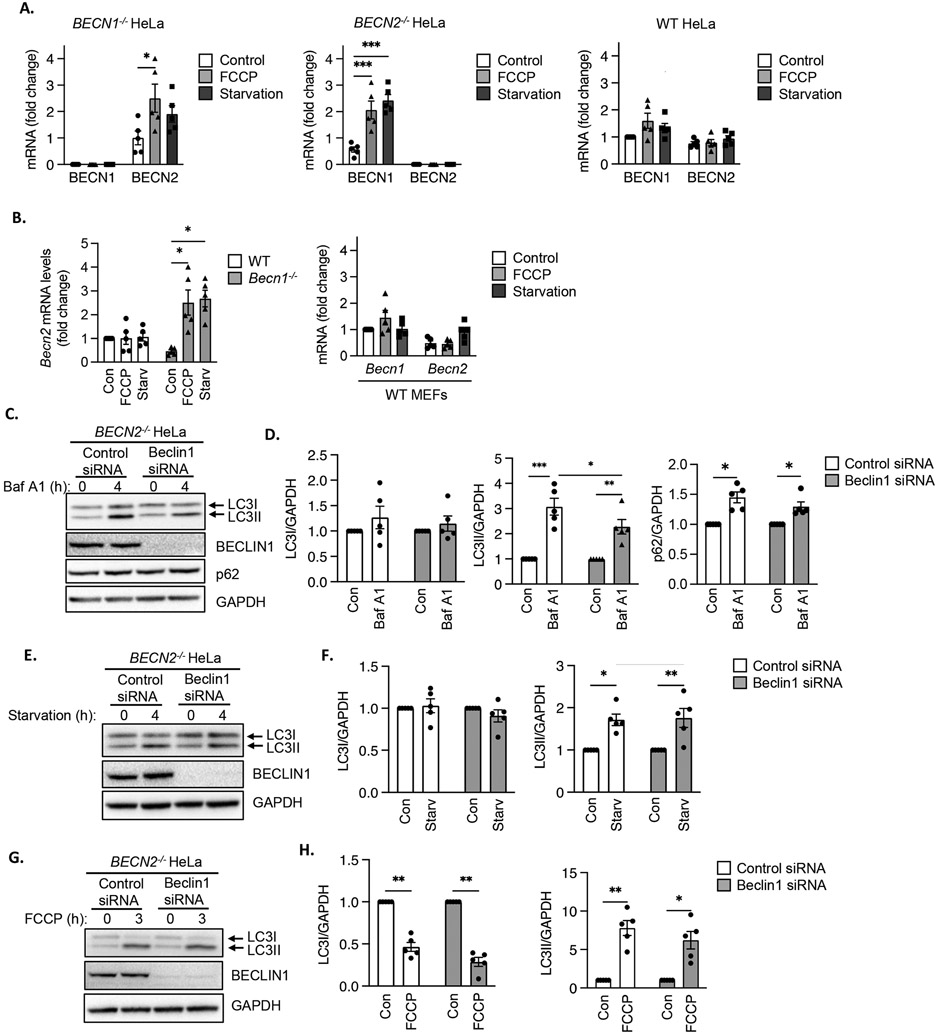
Our findings suggested that neither Beclin1 nor Beclin2 alone was essential for autophagosome formation. Given their homology and reports that both function in autophagy, we investigated whether one homologue could compensate for the other. Transcript levels in BECN1−/− and BECN2−/− HeLa cells were not different from those in WT cells under baseline conditions (fig. S1C). However, compensatory induction of BECN1 and BECN2 transcription was observed in BECN2−/− and BECN1−/− HeLa cells, respectively, following starvation and FCCP treatment (Fig. 3A). In contrast, BECN2 and BECN1 transcript levels did not change in WT HeLa cells under the same conditions. Although BECN1 transcript levels were increased by FCCP treatment or starvation in BECN2−/− HeLa cells, we did not observe a corresponding increase in Beclin1 protein levels (fig.S4A-B), which could be due to rapid protein turnover during autophagy progression. In WT HeLa cells, Beclin1 protein levels declined at 6h and 12h of FCCP treatment (fig. S4A) but were unaffected by starvation (fig. S4B). Transcription in Becn1−/− HeLa cells matched trends observed in WT cells because Becn2 mRNA was also increased following starvation or FCCP treatment whereas Becn1 and Becn2 transcript levels remained unchanged in WT MEFs (Fig. 3B). These findings indicate that Becn1 expression increased in response to the increased demand for autophagosome formation when Beclin2 was absent and vice versa.
Fig. 3. Beclin transcript levels in BECN1−/− and BECN2−/− HeLa cells and effect of Beclin1 silencing on autophagosome formation in BECN2−/− HeLa cells.
A) Quantitative PCR analysis for BECN1 and BECN2 transcript levels in BECN1−/−, BECN2−/− and WT HeLa cells at baseline (Control), after FCCP treatment (4 h) or starvation (3 h) (n=5 biological replicates per group). B) Quantitative PCR analysis for Becn1 and Becn2 transcript levels in WT and Becn1−/− MEFs at baseline or after FCCP treatment or starvation (n=5 biological replicates per group). C) Representative Western blots for LC3, BECLIN1 and p62 in BECN2−/− HeLa cells transfected with control or Beclin1 siRNA. Cells were treated with 100 nM bafilomycin A1 (Baf) for 4 h. D) Quantification of LC3I, LC3II and p62 protein levels. GAPDH was used as a loading control (n=5 independent experiments). E) Representative Western blots for LC3 and BECLIN1 in BECN2−/− HeLa cells transfected with control or Beclin1 siRNA. Cells were subjected to starvation for 4 h. F) Quantification of LC3I and LC3II protein levels (n=5 independent experiments). G) Representative Western blots for LC3 and BECLIN1 in BECN2−/− HeLa cells transfected with control or Beclin1 siRNA. Cells were treated with 25 μM FCCP for 3 h. H) Quantification of LC3I and LC3II protein levels (n=5 independent experiments). Data are presented as means ± SEM. *p<0.05, **p<0.01, ***p<0.001 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test.
We sought to further investigate potential compensation by Beclin1 in BECN2-deficient cells. Silencing Beclin1 in BECN2-deficient HeLa cells led to a small but significant decline in autophagosome formation under baseline conditions as demonstrated by decreased accumulation of LC3II in the presence of Baf A1 (Fig. 3C-D, fig. S12). We did not observe differences in the accumulation of p62, indicating that degradation of autophagic cargo was still intact despite the lack of both Beclin1 and Beclin2 (Fig. 3C-D, fig. S12). However, knockdown of Beclin1 in BECN2−/− HeLa cells did not affect autophagosome formation in response to starvation (Fig. 3E-F) or FCCP treatment (Fig. 3G-H, fig. S12). These findings suggest that although both Beclin1 and Beclin2 might contribute to basal autophagosome formation, they were not essential for stress-induced autophagosome formation.
Beclin1, but not Beclin2, is required for efficient mitophagy
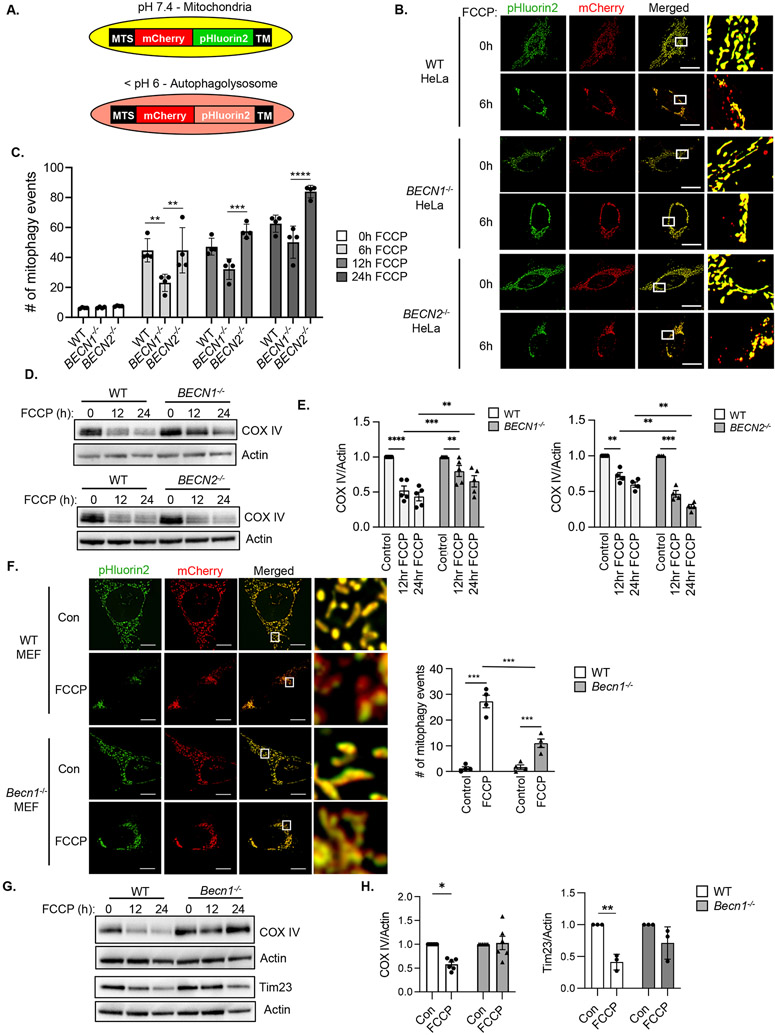
Because autophagosomes are responsible for sequestering damaged mitochondria for delivery to lysosomes during mitophagy (1), we investigated whether mitophagy was altered in BECN1−/− and BECN2−/− HeLa cells. To assess mitophagy, we generated a pH-sensitive dual fluorescent mitophagy reporter consisting of a tandem mCherry-pHluorin2 fluorophore that is targeted to the mitochondrial matrix (Fig. 4A and fig. S6A). Both mCherry (red) and pHluorin2 (green) emission signals are detected under neutral pH, but the pH-sensitive pHluorin2 signal is quenched in the acidic environment of the lysosome whereas the mCherry signal remains intact (16). To investigate the impact of Beclin1 or Beclin2 deficiency on mitophagy, we assessed the number of mitophagy events in cells overexpressing MTS-mCherry-pHluorin2. FCCP treatment increased the number of mitochondria positive only for mCherry fluorescence in all three cell lines, but the number of mitophagy events in BECN1−/− HeLa cells was reduced at 6h compared to WT and BECN2−/− HeLa cells (Fig.4B-C). By 24h, the number of mitophagy events in BECN1−/− HeLa cells had reached levels similar to WT cells, suggesting that the rate of mitophagy was decreased in the absence of Beclin1. The reduced mitophagy in BECN1−/− HeLa cells was also confirmed by Western blotting for the mitochondrial protein COX IV (Fig. 4D-E, fig. S12). Notably, BECN2−/− HeLa cells had increased rates of mitophagy compared to WT and BECN1−/− cells as assessed by mitophagy reporter imaging (Fig. 4B-C) and by Western blot analysis showing accelerated depletion of mitochondrial COX IV levels (Fig. 4D-E). We also confirmed that pharmacological inhibition of Vps34 by SAR405 led to decreased FCCP-mediated clearance of COX IV but did not affect the increase in LC3II (fig. S5). Treatment of cells with oligomycin plus antimycin A for 6 hours to induce mitophagy generated similar results with enhanced mitophagy in BECN2−/− HeLa cells and decreased mitophagy in BECN1−/− cells (fig. S6B-C). Consistent with reduced mitochondrial quality control, maximal mitochondrial respiration was decreased in BECN1−/− but not BECN2−/− HeLa cells (fig. S6D). Finally, we verified that the number of mitophagy events in response to FCCP treatment was also reduced in Becn1−/− MEFs (Fig. 4F). Western blotting confirmed decreased FCCP-mediated clearance of COX IV and Tim23 in Becn1−/− MEFs (Fig. 4G-H, fig. S12). Together, these results suggest that Beclin1, but not Beclin2, functions in mitophagy and that loss of Beclin1 led to a reduced rate of mitochondrial clearance.
Fig. 4. Beclin1-deficient cells have reduced mitophagy.
A) Schematic illustration of the mitophagy reporter which is composed of a mitochondrial targeting sequence (MTS), mCherry (red), pHluorin2 (green) and a transmembrane domain (TM). B) Representative images of WT, BECN1−/− and BECN2−/− HeLa cells overexpressing the mitophagy reporter before and after FCCP (10 μM, 6h) treatment. C) Quantification of the number of mitophagy events per cell after FCCP treatment (n=90 cells/group from 3 experiments). D) Representative Western blots for COX IV in WT, BECN1−/− and BECN2−/− HeLa cells at baseline and after FCCP (25 μM) treatment (n=5 independent experiments). E) Quantification of COX IV protein levels in WT, BECN1−/− and BECN2−/− HeLa cells (n=5 independent experiments). F) Representative images and quantification of the number of mitophagy events per cell in WT and Becn1−/− MEFs overexpressing the mitophagy reporter (n=90 cells/group from 4 experiments). Cells were treated with FCCP (10 μM) for 6 h. G) Representative Western blots for COX IV and Tim23 in WT and Becn1−/− MEFs treated with FCCP (25 μM) for 0, 12 and 24 h. H) Quantification of COX IV and Tim23 protein levels in WT and Becn1−/− MEFs after 12 h of FCCP treatment (n=5 independent experiments). Data are presented as means ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Scale bars = 10 μm.
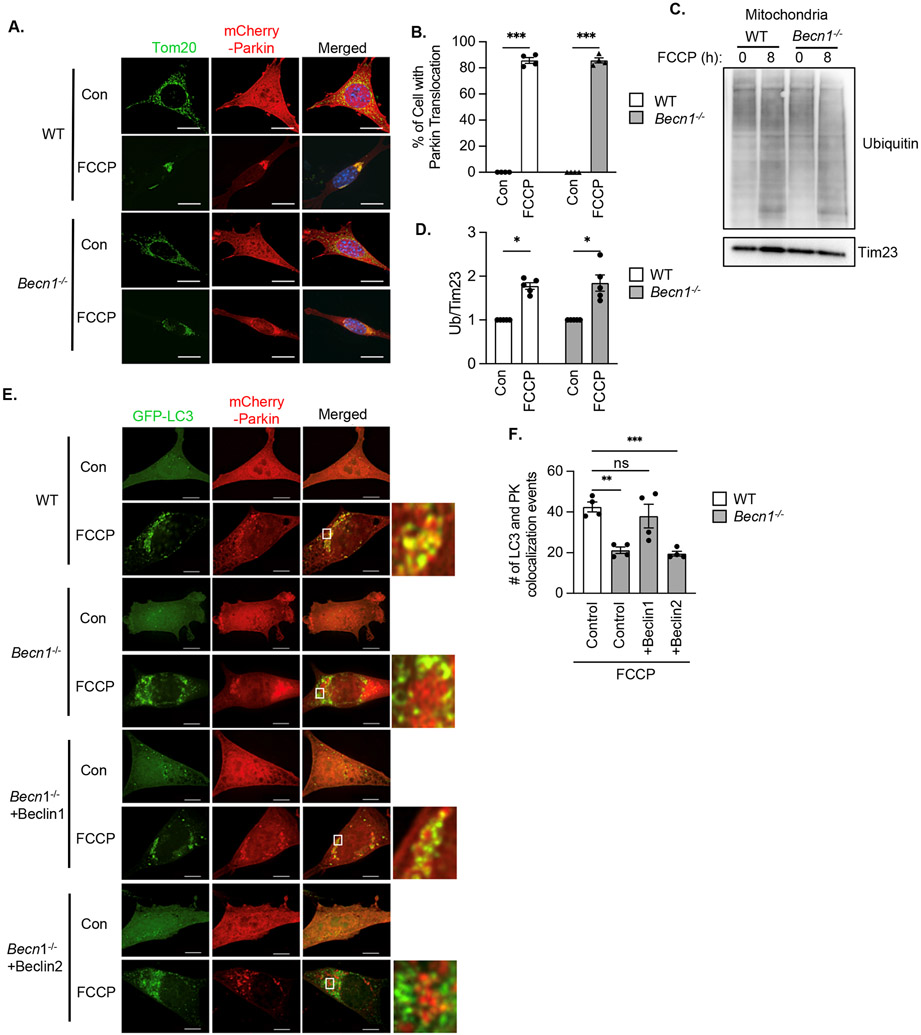
Mitochondria are not sequestered by autophagosomes in the absence of Beclin1
Mitophagy requires two distinct events: labeling of dysfunctional mitochondria by the E3 ubiquitin ligase Parkin for degradation and the formation of autophagosomes near cargo. Upon FCCP treatment, mCherry-Parkin translocation to mitochondria was evident in both WT and Becn1−/− MEFs (Fig. 5A-B). Additionally, ubiquitination of mitochondrial proteins was similarly increased in both WT and Becn1−/− MEFs in response to FCCP (Fig. 5C-D, fig. S12), suggesting that mitochondrial Parkin recruitment occurred independently of Beclin1 Next, we evaluated the colocalization between Parkin-labeled mitochondria and autophagosomes. Although there was increased colocalization between GFP-LC3 positive autophagosomes and mCherry-Parkin in WT MEFs after FCCP treatment, Becn1−/− MEFs had significantly fewer mitochondria that co-localized with autophagosomes under the same conditions (Fig. 5E-F). Overexpression in Becn1−/− MEFs of Beclin1, but not of Beclin2 restored the co-localization between mitochondria and GFP-LC3-labeled autophagosomes (Fig. 5E-F and fig. S7A). Therefore, Beclin1 was necessary for proper targeting of autophagosomes to mitochondria downstream of Parkin translocation whereas Beclin2 was dispensable for mitophagy.
Fig. 5. Parkin translocation in Becn1−/− MEFs.
A) Representative images of WT and Becn1−/− MEFs overexpressing mCherry-Parkin and treated with FCCP (10 μM) for 6h. Cells were stained for Tom20 to label mitochondria and nuclei were counter stained with Hoechst (blue). B) Quantification of the percentage of cells with mitochondrial Parkin after treatment with FCCP (10 μM) for 8h (n=400 cells/group from 4 experiments). C) Representative Western blot for ubiquitin in mitochondrial fractions from WT and Becn1−/− MEFs. D) Quantification of total ubiquitin (Ub) levels (n=5 independent experiments). E) Representative images of WT and Becn1−/− MEFs overexpressing GFP-LC3 and mCherry-Parkin and treated with FCCP (10 μM) for 6h. Cells were analyzed for colocalization between GFP-LC3 puncta and mCherry-Parkin. F) Quantification of GFP-LC3 and mCherry-Parkin co-localization events per cell (n=120 cells/group from 4 experiments). Data are presented as means ± SEM. *p<0.05, **p<0.01, ***p<0.001 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Scale bars = 10 μm.
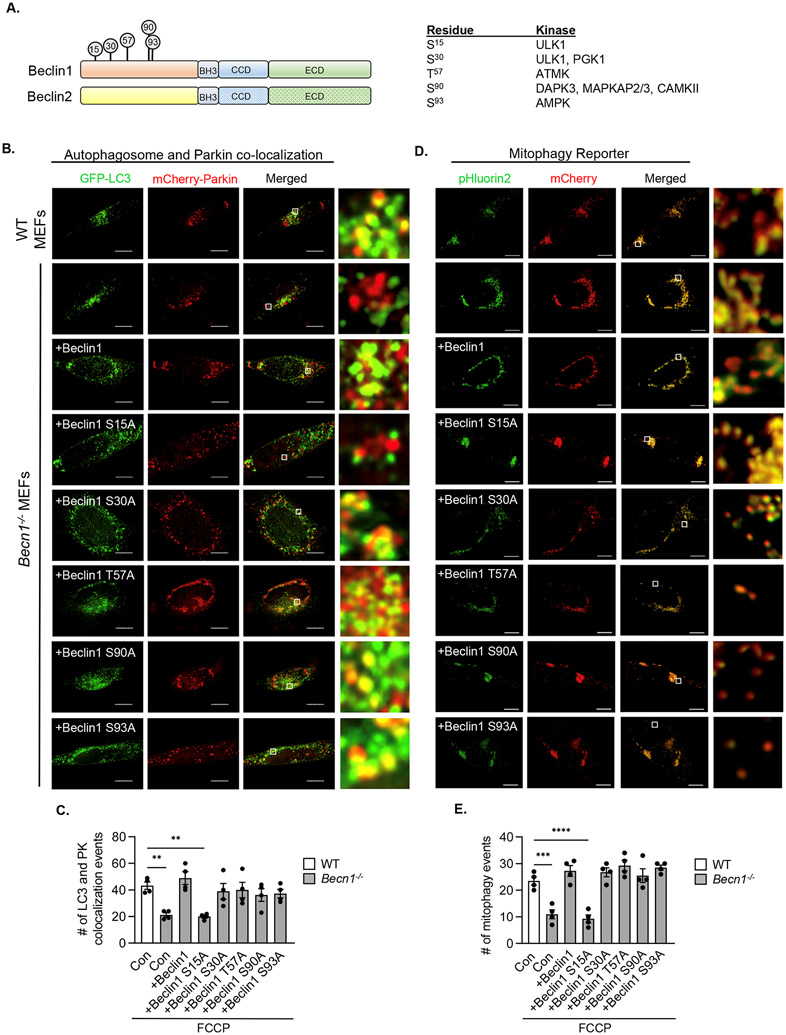
Beclin1 is required for directing autophagosomes to damaged mitochondria during mitophagy
Beclin1 and Beclin2 have distinct N-terminal regions and the phosphorylation of various residues in this region of Beclin1 reportedly affects its function in autophagy (9, 17-24) (Fig. 6A). To examine the potential contribution of these phosphorylation sites in regulating Beclin1’s function in mitophagy, we generated several different phosphorylation-resistant Beclin1 mutants in which Ser15, Ser30, Thr57, Ser90, or Ser93 was changed to alanine (fig. S7B). Overexpression of either Beclin1, Beclin1 S30A, Beclin1 T57A, Beclin1 S90A, or Beclin1 S93A in Becn1−/− MEFs restored the colocalization between GFP-LC3-labeled autophagosomes and mCherry-Parkin positive mitochondria in response to FCCP treatment (Fig. 6B-C). Only Beclin1 S15A failed to restore the co-localization between GFP-LC3 and Parkin-labeled mitochondria (Fig. 6B-C). Experiments using the mitophagy reporter further confirmed the restoration of mitophagy by all mutants except Beclin1 S15A (Fig. 6D-E). We found no changes in the cytosolic distribution of mCherry-Parkin or GFP-LC3 at baseline (fig. S8A-B) or in baseline levels of mitophagy in cells overexpressing the various Beclin1 constructs (fig. S8C-D). Ser15 in Beclin1 can be phosphorylated by Ulk1 (17), a kinase that functions in mitophagy (25), and we found that both starvation and FCCP treatment led to increased phosphorylation of Beclin1 at Ser15 in WT but not Ulk1−/− MEFs (fig. S9A-B). We also confirmed that inhibiting Ulk1 with SBI-0206965 led to reduced mitochondrial clearance in response to FCCP treatment (fig. S10A-B). Because SBI-0206965 can also target AMP-activated kinase (AMPK) (26, 27), another regulator of autophagy and mitophagy (28, 29), we extended these studies to Ulk1−/− MEFs. Similar to Becn1−/− MEFs and WT cells treated with SBI-0206965, Ulkl−/− MEFs showed significantly decreased mitophagy as assessed by Western blotting for COX IV (fig. S10C-D) and in the co-localization between autophagosomes and mCherry-Parkin-labeled mitochondria (fig. S10E-F). Together, these results suggest that Beclin1 ensured proper targeting of autophagosomes to mitochondria through Ulk1-dependent phosphorylation of Ser15.
Fig. 6. Ser15 in Beclin1 and targeting autophagosomes to damaged mitochondria.
A) Protein domains of Beclin1 and Beclin2. Phosphorylation sites in the N-terminal region of Beclin1 and the kinase targeting these sites are indicated. B) Representative images of WT and Becn1−/− MEFs transfected with Beclin1 or Beclin1 phosphorylation resistant mutants plus GFP-LC3 and mCherry-Parkin prior to treatment with 10 μM FCCP for 6h. Cells were stained for HA. White boxes indicate magnified regions. C) Quantification of GFP-LC3 and mCherry-Parkin colocalization events per cell (n=90 cells/group from 3 experiments). D) Representative images of WT and Becn1−/− MEFs overexpressing MTS-mCherry-pHluorin2 plus Beclin1 or Beclin1 mutants and treated with 10 μM FCCP for 12h. White boxes indicate magnified regions. E) Quantification of the number of mitophagy events per cell, as measured by the number of red puncta after subtraction of yellow signal (n=90 cells/group from 3 experiments). Data are presented as means ± SEM. Statistical test: **p<0.01, ***p<0.001, and ****p<0.0001 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Scale bars = 10 μm.
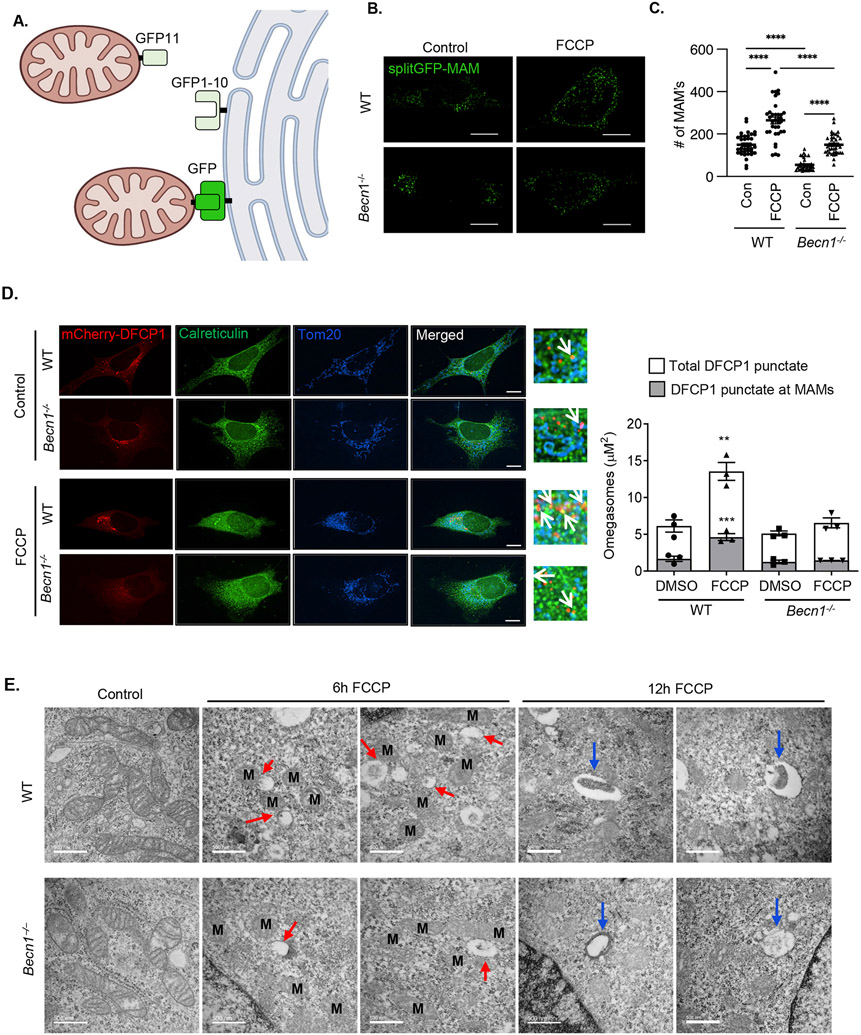
Beclin1 S15A does not associate with MAMs during mitophagy
The fact that autophagosome formation was intact in both Ulk1- and Beclin1-deficient cells suggested that their biogenesis might not be initiated in the proximity of damaged mitochondria. Autophagosome formation can be started at sites known as mitochondria-associated membranes (MAMs) where the endoplasmic reticulum (ER) is physically tethered to mitochondria (30, 31). To investigate the relationship between Beclin1 and MAMs during mitophagy, we utilized a split-GFP reporter to monitor MAM formation in cells (32). In this system, GFP1-10 is anchored in the ER and GFP11 is anchored to the mitochondrion, and when the two GFP fragments are within 10 nm of each other, they form a complete fluorescent GFP protein (Fig. 7A and fig. S11). We found that Becn1−/− MEFs had a reduced number of MAMs at baseline (Fig. 7B-C). The number of MAMs significantly increased in both WT and Becn1−/− MEFs after treatment with FCCP, but to a lesser extent in Becn1−/− MEFs than in WT cells (Fig. 7B-C).
Fig. 7. Phosphorylation at Ser15 and localization of Beclin1 to MAMs during mitophagy.
A) Schematic illustration of the split GFP MAM reporter. B) Representative images of WT and Becn1−/− MEFs overexpressing the MAM reporter and treated with FCCP (10 μM for 6 h). C) Quantification of the average number of MAMs/cell (a total of 30 cells were scored in 5 independent experiments). D) Representative images of WT and Becn1−/− MEFs overexpressing the omegasome reporter mCherry-DFCP1. Cells were treated with FCCP (25 μM) for 4h and stained with anti-Calreticulin and anti-Tom20 to label ER and mitochondria, respectively. Quantification of total mCherry-DFCP1 punctate (white bars) and area colocalized with MAMs (grey bars) (n=90 cells/group from 3 independent experiments). E) Transmission electron microscopy images of WT and Becn1−/− MEFs under baseline conditions and after treatment with 10 μm FCCP. Mitochondria are marked with M, red arrows mark small vesicular structures near mitochondria after FCCP treatment, and blue arrows mark larger vesicles. Scale bar = 500 nm. Data are presented as means ± SEM. **p<0.01, ***p<0.001, and ****p<0.0001 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Scale bars = 10 μm.
Omegasomes are PI3P-enriched ER subdomains that serve as platforms for autophagosome formation and Double FYVE-containing protein 1 (DFCP1) localizes to omegasomes upon induction of autophagy by binding to PI3P through its FYVE domain (33). To investigate whether omegasomes formed at MAMs upon induction of mitophagy, we evaluated the localization of mCherry-DFCP1 in WT and Becn1−/− MEFs. Immunofluorescence analysis confirmed a significant increase in overall omegasome formation in response to FCCP treatment in WT MEFs (Fig. 7D). Moreover, we found that WT MEFs, but not Becn1−/− MEFs, had increased mCherry-DFCP1 co-localization to areas of overlap between mitochondria (marked by Tom20 fluorescence) and ER (marked by Calreticulin fluorescence), suggesting increased omegasome formation at MAMs during mitophagy. Transmission electron microscopy analysis demonstrated formation of small vesicular structures in close proximity to mitochondria in WT MEFs after 6h of FCCP treatment and engulfed material enclosed in vesicles by 12h (Fig. 7E). In contrast, Becn1−/− MEFs appeared to have fewer membrane structures adjacent to damaged mitochondria at 6h and empty vesicular structures were present in cytosol at 12h of FCCP treatment.
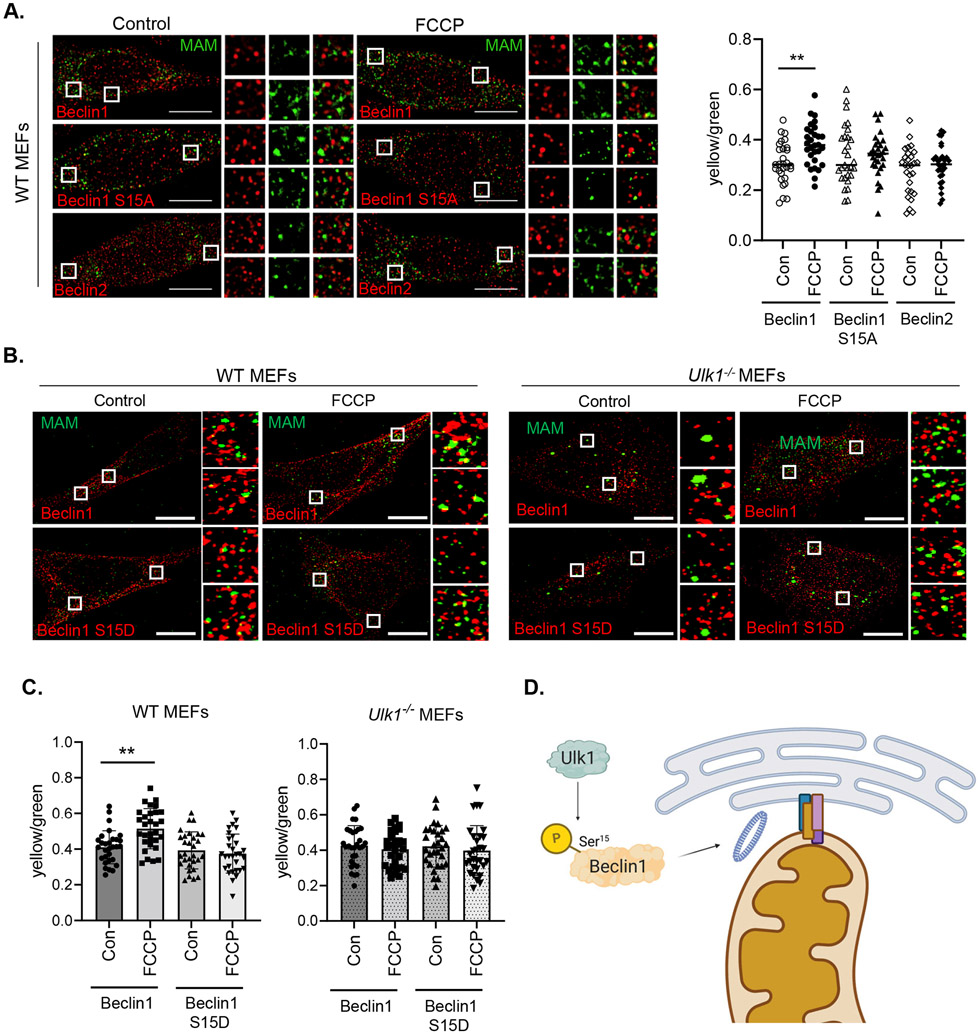
Finally, using our slit-GFP reporter system, we confirmed that induction of mitophagy by FCCP led to increased association of Beclin1 with MAMs (Fig. 8A). In contrast, neither Beclin1 S15A nor Beclin2 showed increased co-localization with MAMs upon induction of mitophagy. We also found that the association between Beclin1 and MAMs did not increase in Ulk1−/− MEFs after FCCP treatment (Fig. 8B-C), suggesting that Ulk1-mediated phosphorylation of Ser15 in Beclin1 was critical for autophagosome formation at MAMs during mitophagy induction. The phosphomimetic Beclin1 S15D mutant did not show increased localization to MAMs in WT and Ulk1−/− MEFs at baseline or after FCCP treatment. Overall, these findings suggest that Ulk1-mediated recruitment of Beclin1 to MAMs is necessary for the effective initiation of autophagosome formation in the proximity of damaged mitochondria upon induction of mitophagy.
Fig. 8. Beclin1 and MAM localization in WT and Ulk1−/− MEFs.
A) Representative images of WT MEFs overexpressing the split-GFP MAM reporter plus Beclin1, Beclin1 S15A, or Beclin2. Cells were treated with DMSO or 10 μM FCCP for 6h. The fluorescence intensity of co-localization (yellow) signal normalized to total MAMs (green) was quantified in cells (n=30 cells/group from three independent experiments). B) Representative images of WT and Ulk1−/− MEFs overexpressing Beclin1 or Beclin1S15D plus the split-GFP MAM reporter. Cells were treated with 10 μM FCCP for 6h. C) Quantification of Beclin1-MAM colocalization events/total MAMs on a per cell basis (n=30 cells/group from three independent experiments). D) Proposed model illustrating role of Ulk1-mediated phosphorylation of Beclin1 at Ser15 during mitophagy. Phosphorylated Beclin1 re-localizes to a mitochondria-ER associated membrane site where it initiates formation of the autophagosome membrane adjacent to a dysfunctional mitochondrion. Data are presented as means ± SEM. **p<0.01 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Scale bars = 10 μm.
DISCUSSION
The findings in this study provide important new insights into the functions of Beclin1 and Beclin2 in autophagy and mitophagy. First, our data demonstrated that both Beclin1 and Beclin2 contributed to formation of LC3II-positive vesicles under baseline conditions, whereas a Beclin1/2-independent mechanism of autophagosome formation was activated in response to stress. In addition, our findings established that Beclin1, but not Beclin2, functioned in mitophagy, and that Ulk1-mediated phosphorylation of Ser15 in Beclin1’s distinct N-terminal region was required for its association with MAMs during mitophagy. We propose that the localization of Beclin1 at MAMs ensures that autophagosome formation is initiated adjacent to damaged mitochondria, leading to their efficient engulfment (Fig. 8D).
Our data from both mouse and human cells provide genetic evidence for a Beclin1/2-independent autophagy pathway that is activated by distinct stressors. Although the formation of autophagosomes under baseline conditions involved Beclin1 and Beclin2, the absence of these proteins did not prevent an increase in LC3II-positive vesicle formation in response to starvation or FCCP treatment, thus confirming the existence of a Beclin1/2-independent pathway that can initiate autophagy under these conditions. The finding that the autophagy adaptor protein p62 did not accumulate in Beclin1/2-deficient cells further confirmed intact intracellular degradation of autophagic cargo. Accumulation of p62 in cells is an indicator of impaired autophagic degradation (34, 35). Studies have identified a Beclin1-Vps34 independent autophagy pathway for which genetic deletion or silencing of Beclin1 or Vps34 has little effect on autophagosome formation in response to different treatments (36-39). Beclin1 and Beclin2 are core components of the Class III PI3K complex that generates PI3P, the key phospholipid responsible for recruiting downstream effector proteins involved in autophagosome formation and elongation (40). We found that Beclin1 deficiency led to a reduced increase in localized PI3P generation in response to FCCP treatment compared to WT cells. This result suggests the specific activation of the Beclin1-Vps34 complex during mitophagy and that Beclin2 is not compensating for Beclin1 in the PI3K complex during mitophagy. Moreover, it has been reported that PI5P can recruit WIPI2 and DFCP1 effector proteins to induce autophagosome formation in the absence of Vps34 and PI3P (41). However, FCCP treatment failed to recruit DFCP1 to MAMs in Beclin1 deficient cells suggesting that PI5P is not compensating in the formation of LC3II-positive vesicles. It also raises the important question of how autophagosomes are formed in the absence of Beclin1 Various modulators of autophagy, including mitochondrial uncouplers, have been reported to activate a parallel non-canonical autophagy pathway that is independent of PI3P generation but that involves conjugation of LC3II to endolysosomal membranes (42). Flence, it is possible that that the LC3II-positive vesicles in Beclin1 deficient cells represents activation of a distinct non-canonical autophagy.
The function of Beclin1 in mitophagy is still unclear due to conflicting findings (6-8). Here, we discovered that mitophagy was decreased in the absence of Beclin1 despite an increased number of autophagosomes upon induction of mitophagy. Beclin1 has been reported to interact with Parkin to facilitate its translocation to mitochondria during mitophagy (6). However, our studies revealed that Parkin recruitment and ubiquitination of mitochondrial proteins were intact in Beclin1-deficient cells, suggesting that reduced labeling of mitochondria did not account for the decreased mitophagy. The reasons for these different findings are unclear. It is possible that a portion of Parkin interacts with the Beclin1-Vps34 core complex to ensure their concurrent delivery to dysfunctional mitochondria upon activation of mitophagy.
Unexpectedly, the rate of mitochondrial clearance was increased in Beclin2-deficient cells. The reason for this finding is unclear but it is possible that depolarized mitochondria might be delivered to lysosomes through both traditional autophagosomes as well as alternative autophagosomes or early endosomes. Autophagosomes formed through an alternative Atg5/7-independent, Rab9-dependent pathway can engulf mitochondria for delivery to lysosomes (43, 44). Similarly, we have previously reported that Rab5-positive early endosomes can engulf dysfunctional mitochondria for lysosomal degradation (35, 45).
Beclin1 shares about 57% sequence homology with Beclin2 (9) and shares several domains that are essential for interactions with proteins in the PI3K complex (6-8). The N-terminal regions in Beclin1 and Beclin2 are highly divergent, and our findings demonstrated that this domain was responsible for Beclin1’s function in mitophagy. This region contains several conserved phosphorylation sites that can modulate its function. For instance, Russel et al. discovered that amino acid deprivation triggers the Ulk1-dependent phosphorylation of Beclin1 at Ser15, which activates the PI3K complex and autophagy (17). A different study reported that Ulk1-mediated phosphorylation at Ser30 is associated with increased PI3K activity and autophagosome formation in response to starvation or hypoxia (19). We uncovered that Ser15 was important for its function in mitophagy, whereas phosphorylation of Ser30 is not required for this function. Thus, phosphorylation of Beclin1 at specific residues might dictate whether it functions in selective mitophagy versus general autophagy.
MAMs provide a platform for efficient crosstalk between the ER and mitochondria and autophagosome formation at MAMs facilitates the biogenesis of the autophagosome proximal to a dysfunctional mitochondrion (31). Beclin1 re-localizes to MAMs upon induction of mitophagy (7) and here, we provided new evidence that Ulk1-mediated phosphorylation of Beclin1 at Ser15 was needed for its translocation to MAMs. Beclin2 lacks the Ulk1 phosphorylation site that is present in Beclin1 and did not localize to MAMs during mitophagy. These findings further confirm that the unique N-terminal region in Beclin1 is responsible for its distinct function in mitophagy. Unexpectedly, the S15D phosphomimetic Beclin1 mutant did not localize to MAMs at baseline or after activation of mitophagy in WT and Ulk1−/− MEFs. This finding suggests that phosphorylation of Ser15 alone might not be sufficient to promote its translocation to MAMs and that constitutive phosphorylation of the residue might even inhibit this process. Also, Ulk1-dependent phosphorylation of Ser15 in Beclin1 was observed in response to both starvation and FCCP treatment, suggesting that Beclin1 is recruited to MAMs to initiate autophagosome formation during both general and selective autophagy. Multiple autophagosome biogenesis sites exist at the ER that are utilized during general autophagy. However, localized autophagosome formation at MAMs is more critical during mitophagy. Moreover, hearts in Beclin1 hemizygous mice have been reported to exhibit fewer MAMs (46), suggesting that Beclin1 is involved in MAM formation or stability. Similarly, we found that the number of MAMs were reduced in Becn1−/− MEFs at baseline and after induction of mitophagy compared to WT MEFs. Along these lines, fewer sites for initiating autophagosome formation in the proximity of dysfunctional mitochondria will lead to less efficient engulfment of cargo.
In summary, our study has uncovered that Beclin1 and Beclin2 are dispensable for autophagosome formation in cells and that a stress-responsive Beclin1/2-independent mechanism exists. We also discovered that Beclin1 plays a specific role in mitophagy and that Ulk1 is an upstream regulator of the re-localization of Beclin1 to MAMs during mitophagy. Overall, we propose that Ulk1-mediated phosphorylation of Beclin1 at Ser15 during mitophagy facilitates its translocation to MAMs for autophagosome initiation proximal to damaged mitochondria that need to be eliminated. Future studies should focus on identifying the Beclin1/Beclin2-independent mechanism of autophagy that is activated during stress and the relationship between MAMs and Beclin1 in mitophagy.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
Cell lines were cultured in DMEM+ Glutamax media (GIBCO) supplemented with 10% FBS (GIBCO) and 1% antibiotic-antimycotic (GIBCO) and maintained in 5% CO2 at 37°C. All cells were routinely tested for mycoplasma. Wild-type (WT) and Becn1−/− mouse embryonic fibroblasts (MEFs) were generously gifted by Dr. Zhenyu Yue (Icahn School of Medicine at Mount Sinai) (47). WT and Ulk1−/− MEFs were generously provided by Dr. Mondira Kundu (St. Jude’s Research Hospital) (25). BECN1−/− and BECN2−/− HeLa cells were generated using the Gene Knockout Kit v2 from Synthego according to the manufacturer’s protocol. Briefly, Cas9 2NLS nuclease was combined with Beclin1 or Beclin2 multi-gRNA oligo mixture (Beclin1 gRNAs: 5' CUCGCUGACCUGUGAGUUCC 3', 5' UCGUGUCCAGUUUCAGGGGC 3' and 5' GUCCAACAACAGCACCAUGC 3'; Beclin2 gRNAs: 5' GCGCUGGCACAGGAAGCGGA 3', 5' GCAGCCCCGGCGCCCACCUC 3' and 5' CACCUCCCUGGUGGUGACGC 3') in Nucleofector solution to form ribonucleoprotein (RNP) complexes. RNP complexes were electroporated into HeLa cells using a Lonza 4D-Nucleofector. Cells were plated for 72 h in culture media before being plated for single-cell clonal expansion. Gene deletion was validated using Sanger sequencing, the ICE v2 analysis tool, qPCR and Western blotting.
Plasmid Constructs
GFP-LC3 (plasmid 11546), Myc-Parkin (plasmid 17612), mCherry-Parkin (plasmid #23956), pcDNA4-Beclin1-HA (plasmid #24399) (48), GFP-p40PX (plasmid #19010) and mCherry-DFCP1 (plasmid 86746) were obtained from Addgene. HA-Beclin1 S15A, HA-Beclin1 S15D, HA-Beclin1 S30A, HA-Beclin1 T57A, HA-Beclin1 S90A, HA-Beclin1 S93A mutants and HA-Beclin2 constructs were synthesized by Genscript using pcDNA3.1(+) as a backbone. The mitophagy reporter and split GFP-MAM reporter constructs were synthesized by Genscript. The mitophagy reporter MTS-mCherry-pHLuorin2-TM contains the tandem mCherry-pHleurin2 fused to the mitochondrial targeting sequence (MTS) from ATP synthase and the transmembrane (TM) domain from human Myeloid cell leukemia 1. For the split-GFP-MAM reporter, GFP1-10 was targeted to the endoplasmic reticulum with residues 228-259 from ubiquitin-conjugating enzyme E2 J2 (V5-ER-GFP1-10), and GFP11 was targeted to mitochondria with residues 1-59 from translocase of the outer mitochondrial membrane 70 protein (myc-mito-2XGFP11) as previously described (32).
Antibodies
The following antibodies were used for immunofluorescence (IF) and Western blot (WB) experiments: anti-Beclin1 (IF 1:100, WB 1:1000; Santa Cruz, sc-11427), anti-Actin (WB 1:1000; Genetex, 110003), anti-GAPDH (WB 1:1000; GenTex, GTX627408), anti-p62 (WB 1:1000; Abcam, ab56416), anti-LC3 A/B (WB 1:1000; Cell Signaling, 4108), anti-MTC01 (labeled as COX IV; IF 1:100, WB 1:1000; Thermo, 459600), anti-Parkin (WB 1:1000; Cell Signaling, 4211), anti-Tim23 (IF 1:100; WB 1:1000; BD Biosciences, 611222), anti-Tom20 (IF 1:100, WB 1:1000; Santa Cruz, 11415, and IF 1:100;Abcam, ab56783), anti-Ubiquitin (WB 1:1000; Enzo, ABS840), anti-Tubulin (WB 1:1000; Sigma, T6074), anti-Myc (IF 1:100; WB 1:1000; Sigma, M4439), anti-Flag (IF 1:100; WB 1:1000; Sigma, F7425), anti-Calreticulin (IF 1:100; Abcam, ab2907) anti-Cytochrome C (IF 1:100; BD Pharmigen, 556432) and anti-HA (IF 1:100; Cell Signaling 37245). Secondary antibodies used for WB were goat anti-mouse FIRP (Life Technologies, 31430) and goat-anti-rabbit HRP (Life Technologies, 31460). Secondary antibodies used for IF were Alexa Fluor 350 (Thermo Fisher, rb A11046, ms A11045), Alexa Fluor 488 (Thermo Fisher, rb A11034, ms A11029), Alexa Fluor 594 (Thermo Fisher, rb A11037, ms A11032), and Alexa Fluor 647 (Thermo Fisher, rb A21244, ms A21235).
Transient transfections and adenoviral infections
DNA transfections were performed using Fugene 6 Transfection Reagent (Promega, E269A). siRNA transfections were done using RNAiMax lipofectamine reagent (Thermo Fisher, 13778) according to the manufacturer’s protocol. For mitophagy experiments, cells were transfected or infected with Myc-Parkin or Flag-Parkin. All experiments were performed 24 h post-transfection unless otherwise noted. BECN2−/− HeLa cells were transiently transfected with 50nM of negative control siRNA (Qiagen, SI03650318; seq: 5' CAGGGTATCGACGATTACAAA3') or 50nM human Beclin1 siRNA (Sigma siRNA ID: SASI_Hs01_00090913). Cells were infected with the indicated adenovirus in DMEM+ Glutamax with 2% heat-inactivated serum and experiments were performed 24 h later. Adenoviruses mCherry-Parkin (MOI 50) and GFP-LC3 (MOI 150) have been previously described (49). MTS-mcherry-pHluorin2-TM (MOI 50) and Flag-Parkin (MOI 50) were generated by the viral core facility at the University of California, Davis.
Western Blot Analysis
Cell lysates were prepared by homogenizing cells in cold lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100) and complete protease inhibitor cocktail (Roche Applied Science, 11873580001). Lysates were cleared by centrifugation at 20,000 x g for 20 minutes at 4°C and protein concentrations were assessed by the Bradford assay. Mitochondrial fractions were isolated from MEFs as described (50). Briefly, cells were homogenized in ice cold isolation buffer containing 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 10 mM HEPES, and protease inhibitors using a plastic Eppendorf tube mortar and pestle on ice to gently release mitochondria. This suspension was centrifuged at 1000 x g for 5 minutes at 4°C to pellet cellular debris. The supernatant was spun at 14,000 x g at 4°C for 15 minutes and the mitochondrial pellet was gently washed and resuspended in isolation buffer. Proteins were separated on NuPAGE Bis-Tris gels (Invitrogen, NP0302BOX) and transferred to nitrocellulose membranes (GE Healthcare Life Sciences, 10600001). After blocking in 5% milk TBS-Tween, membranes were incubated with the specified antibodies at 4° C overnight, followed by secondary antibody for 1 hour. Bands were developed using Bio-Rad ChemiDoc XRS+ imager and densitometry quantification was performed using Image Lab 4.1 (Bio-Rad) software.
Quantitative PCR
RNA was extracted from cells using the RNeasy Mini Kit (Qiagen, 74104) and quantified by Nanodrop (Thermo Fisher). Reverse transcription to cDNA was performed with the QuantiTect Reverse Transcription Kit (Qiagen, 205311) according to the manufacturer’s protocol. qPCR was performed with standard TaqMan primers and TaqMan Universal Mastermix II (Applied Biosystems 4440040) or SYBR Green (Applied Biosystems, 4309155) on a CFX96 real-time PCR detection system (Bio-Rad Laboratories). Fold difference was calculated by the comparative CT(2-ΔΔCT) method against 18s ribosomal RNA (ThermoFisher, 18S Mm03928990) (51). Becn1 primers were designed to recognize exon2 (forward: 5’- GCATGGAGGGGTCTAAGGCGTC-3’ and reverse: 5’- GTTCCTGGATGGTGACCCGGTC-3’) and were obtained from Eton Bioscience Inc. Becn2 primers (forward: 5’- GCGCTCTCGAGGCTAGCATGTCTTCCATCC-3’ and reverse: 5’- GCGCTAAGCTTGGGCCCTCAAGCGTA-3’) were obtained from Integrated DNA Technologies.
Immunofluorescence and Analysis
Cells were fixed with 4% formaldehyde solution (Thermo Fisher, 28906) in phosphate-buffered saline (PBS), permeabilized with 0.2% Triton X-100 (Sigma, T9284), and blocked in 5% normal goat serum (Vector labs, S-1000). Cells were stained with primary antibodies overnight at 4°C, washed with PBS, and incubated with the secondary antibodies for 1 hour at 37°C. Fluorescence images were captured using a Nikon Eclipse Ti2-E with a motorized XYZ-stage fitted with a Plan-Apochromat lambda 60X or 100X NA 1.40 oil immersion objective. Z stacks were separated by 0.3 μM and acquired with a DS-Qi2 camera (Nikon) illuminated by a Solid-state White Light Excitation Source (Lumencor). Images were processed by 3D deconvolution and compressed into extended depth of focus (EDF) images by NIS-Elements AR GA3 software. Image analysis was performed using NIS elements software and Image J. Colocalization events were quantified by counting puncta that displayed both red and green signal or by Pearson’s correlation coefficient. Mitophagy events using the mitophagy reporter were quantified as red-only puncta that were 2-fold brighter than background and yellow puncta were intentionally excluded. For omegasome analysis, mCherry-DFCP1 puncta above background were identified and their colocalization with MAMs was measured by quantifying the area of mCherry puncta positive for both mitochondria (anti-Tom20 or anti-Cytochrome c immunofluorescence) and endoplasmic reticulum (anti-Calreticulin IF) on a per cell basis. In split-GFP experiments, MAM formation was assessed and quantified with NIS Elements software on single Z-stack slices by formation of GFP-positive structures 1.25-fold above background.
Transmission Electron Microscopy
WT and Becn1−/− MEFs were infected with adenoviruses encoding Myc-tagged Parkin (Ad-Myc-Parkin) at 50 multiplicity of infection (MOI) for 24h prior to 10μM FCCP treatment for 6 or 12h. Cells were fixed with 2% glutaraldehyde in 0.1M sodium cacodylate (SC) buffer at pH7.4 for 1 hour at 4°C. After 5 washes with 0.1M SC, cells were incubated for 30 min with 1% tetroxide osmium in 0.1 M SC buffer on ice, washed again with SC buffer and rinsed with ddH2O. Next, cells were dehydrated for 1min using 20%, 50%, 70%, 90%, and 2x 100% ethanol and incubated 2x1min with acetone. For epoxy resin embedding, cells were incubated 3x1h with durcupan and left to cure for 36 h at room temperature. 60nm ultrathin sections were cut with diamond knife and mounted on 300 mesh grids. Grids were post stained with 2% of uranyl acetate for 5min and Sato’s lead (1% lead acetate, 1% Lead Nitrate, 1% Lead citrate and 2% sodium citrate in water) for 1min. Images were acquired with a TEM Jeol 1400 plus TEM operated at 80 KeV and equipped with a bottom-mounted Gatan One View camera.
Mitochondrial Respiration
Mitochondrial oxygen consumption of HeLa cells was measured using the Seahorse XFp extracellular flux analyzer (Agilent) as previously described (52). Briefly, 2.5x 104 cells per well were plated 24 hours before measurement of oxygen consumption rates. The next day, culture medium was changed to XF Assay Medium (Agilent) containing in DMEM containing 10 mM glucose, 3 mM glutamine, and 1 mM pyruvate. Basal respiration was measured prior to the addition of oligomycin (1μM) to measure ATP-linked respiration. Next, three successive additions of 1μM carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP) were added to measure maximal respiration. Finally, 1 μM Antimycin A was added to measure non-mitochondrial oxygen consumption. Data was analyzed using GraphPad Prism software (version 6.0c) and normalized to cell count using the Cyquant nuclear stain (Invitrogen).
Statistical Analysis
All experiments were independently repeated in biological replicates. Data are expressed as mean ± S.E.M. Differences between groups were assessed by repeated-measure ANOVA tests with Tukey’s post hoc test using GraphPad Prism 6 software. Each test was used based on the number of samples being analyzed according to the test’s assumptions and requirements. Differences were considered significant when p<0.05. Analyses were done unblinded with respect to sample identity. Scientific justifications for data exclusion included very low transfection efficiency (<15%) or changes in cell viability under control conditions.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
Supplementary Material
Acknowledgments
Illustrations were created with BioRender.com.
Funding:
National Institutes of Health grant R01HL155281 (ÅBG)
National Institutes of Health grant R01HL157265 (ÅBG)
National Institutes of Health grant T32GM007752 (MAL)
American Heart Association Postdoctoral Fellowship (JMQ)
Footnotes
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Gustafsson AB, Dorn GW 2nd, Evolving and Expanding the Roles of Mitophagy as a Homeostatic and Pathogenic Process. Physiol Rev 99, 853–892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Autophagy in Human Diseases. N Engl J Med 383, 1564–1576 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Kang R, Zeh HJ, Lotze MT, Tang D, The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18, 571–580 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsunaga K et al. , Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11, 385–396 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Burman JL et al. , Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J Cell Biol 216, 3231–3247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choubey V et al. , BECN1 is involved in the initiation of mitophagy: it facilitates PARK2 translocation to mitochondria. Autophagy 10, 1105–1119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelmetti V et al. , PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 13, 654–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollville E, Carroll RG, Cullen SP, Martin SJ, Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell 55, 451–466 (2014). [DOI] [PubMed] [Google Scholar]
- 9.He C et al. , Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell 154, 1085–1099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu M et al. , Beclin 2 negatively regulates innate immune signaling and tumor development. J Clin Invest 130, 5349–5369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue Z, Jin S, Yang C, Levine AJ, Heintz N, Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc.Natl.Acad.Sci.U.S.A 100, 15077–15082 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill SM, Wrobel L, Rubinsztein DC, Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation. Cell Death Differ 26, 617–629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky DJ et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy 17, 1–382 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lampert MA et al. , BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nascimbeni AC, Codogno P, Morel E, Local detection of Ptdlns3P at autophagosome biogenesis membrane platforms. Autophagy 13, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahon MJ, pHluorin2: an enhanced, ratiometric, pH-sensitive green florescent protein. Adv Biosci Biotechnol 2, 132–137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell RC et al. , ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15, 741–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X et al. , Phosphoglycerate Kinase 1 Phosphorylates Beclin1 to Induce Autophagy. Mol Cell 65, 917–931 e916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JM et al. , ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 14, 584–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang N, Liu X, Zhang S, Sun H, The role of Beclin 1 in IR-induced crosstalk between autophagy and G2/M cell cycle arrest. Cell Signal 62, 109353 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Wei Y et al. , The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X et al. , CaMKII-mediated Beclin 1 phosphorylation regulates autophagy that promotes degradation of Id and neuroblastoma cell differentiation. Nat Commun 8, 1159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujiwara N, Usui T, Ohama T, Sato K, Regulation of Beclin 1 Protein Phosphorylation and Autophagy by Protein Phosphatase 2A (PP2A) and Death-associated Protein Kinase 3 (DAPK3). J Biol Chem 291, 10858–10866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J et al. , Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundu M et al. , Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112, 1493–1502 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahwazi D et al. , Investigation of the specificity and mechanism of action of the ULK1/AMPK inhibitor SBI-0206965. Biochem J 478, 2977–2997 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dite TA et al. , AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965. J Biol Chem 293, 8874–8885 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan DF et al. , Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Kundu M, Viollet B, Guan KL, AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamasaki M et al. , Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Zachari M et al. , Selective Autophagy of Mitochondria on a Ubiquitin-Endoplasmic-Reticulum Platform. Dev Cell 50, 627–643 e625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cali T et al. , splitGFP Technology Reveals Dose-Dependent ER-Mitochondria Interface Modulation by alpha-Synuclein A53T and A30P Mutants. Cells 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Axe EL et al. , Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182, 685–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N, Yoshimori T, Flow to interpret LC3 immunoblotting. Autophagy 3, 542–545 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Hammerling BC et al. , A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat Commun 8, 14050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu JH et al. , Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol 170, 75–86 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grishchuk Y, Ginet V, Truttmann AC, Clarke PG, Puyal J, Beclin 1-independent autophagy contributes to apoptosis in cortical neurons. Autophagy 7, 1115–1131 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R, Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ 15, 1318–1329 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Willinger T, Flavell RA, Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A 109, 8670–8675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bilanges B, Posor Y, Vanhaesebroeck B, PI3K isoforms in cell signalling and vesicle trafficking. Nat Rev Mol Cell Biol 20, 515–534 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Vicinanza M et al. , PI(5)P regulates autophagosome biogenesis. Mol Cell 57, 219–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacquin E et al. , Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy 13, 854–867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishida Y et al. , Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Saito T et al. , An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Invest 129, 802–819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammerling BC, Shires SE, Leon LJ, Cortez MQ, Gustafsson AB, Isolation of Rab5-positive endosomes reveals a new mitochondrial degradation pathway utilized by BNIP3 and Parkin. Small GTPases, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, Cai Y, Qian S, Chiou H, Zang QS, Beclin-1 improves mitochondria-associated membranes in the heart during endotoxemia. FASEB Bioadv, 123–135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKnight NC et al. , Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet 10, e1004626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Q et al. , Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 105, 19211–19216 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanna RA et al. , Microtubule-Associated Protein 1 Light Chain 3 (LC3) Interacts with Bnip3 Protein to Selectively Remove Endoplasmic Reticulum and Mitochondria via Autophagy. J Biol Chem 287, 19094–19104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shires SE et al. , Nuclear Parkin Activates the ERRalpha Transcriptional Program and Drives Widespread Changes in Gene Expression Following Hypoxia. Sci Rep 10, 8499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative C(T) method. Nature protocols 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Orogo AM et al. , Accumulation of Mitochondrial DNA Mutations Disrupts Cardiac Progenitor Cell Function and Reduces Survival. J Biol Chem 290, 22061–22075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.