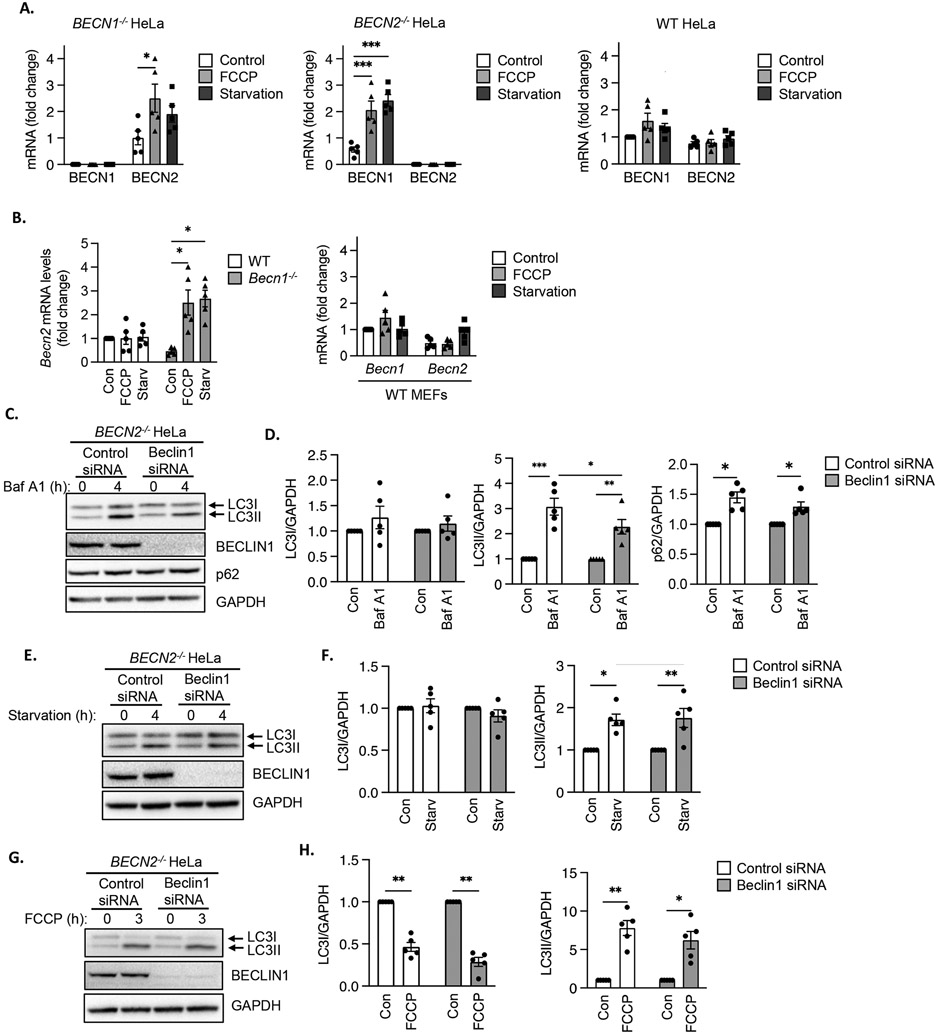
Fig. 3. Beclin transcript levels in BECN1−/− and BECN2−/− HeLa cells and effect of Beclin1 silencing on autophagosome formation in BECN2−/− HeLa cells.
A) Quantitative PCR analysis for BECN1 and BECN2 transcript levels in BECN1−/−, BECN2−/− and WT HeLa cells at baseline (Control), after FCCP treatment (4 h) or starvation (3 h) (n=5 biological replicates per group). B) Quantitative PCR analysis for Becn1 and Becn2 transcript levels in WT and Becn1−/− MEFs at baseline or after FCCP treatment or starvation (n=5 biological replicates per group). C) Representative Western blots for LC3, BECLIN1 and p62 in BECN2−/− HeLa cells transfected with control or Beclin1 siRNA. Cells were treated with 100 nM bafilomycin A1 (Baf) for 4 h. D) Quantification of LC3I, LC3II and p62 protein levels. GAPDH was used as a loading control (n=5 independent experiments). E) Representative Western blots for LC3 and BECLIN1 in BECN2−/− HeLa cells transfected with control or Beclin1 siRNA. Cells were subjected to starvation for 4 h. F) Quantification of LC3I and LC3II protein levels (n=5 independent experiments). G) Representative Western blots for LC3 and BECLIN1 in BECN2−/− HeLa cells transfected with control or Beclin1 siRNA. Cells were treated with 25 μM FCCP for 3 h. H) Quantification of LC3I and LC3II protein levels (n=5 independent experiments). Data are presented as means ± SEM. *p<0.05, **p<0.01, ***p<0.001 by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test.