Abstract
Leaf rust caused by Puccinia triticina Erikss. (Pt) is the most widely distributed and important wheat disease worldwide. The objective of the present study was to determine the frequency of Iranian Pt races, their virulence to key resistance genes and map quantitative trait loci (QTL) for resistance to different Pt races from 185 globally diverse wheat genotypes using a genome-wide association study (GWAS) approach. The virulence pattern of the 33 Pt isolates from various wheat-growing areas of Iran on 55 wheat differentials showed that the FKTPS and FKTTS were relatively frequent pathotypes among the 18 identified races. The weighted average frequency of virulence on the resistance genes Lrb, Lr3bg, Lr14b, Lr16, Lr24, Lr3ka, Lr11 and Lr20 were high (> 90%). However, low virulence on the resistant genes Lr2a, Lr9, Lr19, Lr25, Lr28 and Lr29 indicates that these genes are still effective against the pathogen population in Iran at present. GWAS on a panel of 185 wheat genotypes against 10 Pt races resulted into 62 significant marker-trait associations (MTAs) belonged to 34 quantitative trait loci (QTL) across 16 chromosomes. Among them, 10 QTLs on chromosomes 1A, 1B, 3B, 3D, 4A, 6D, 7A and 7D were identified as potential novel QTLs, of which four QTLs (QLr.iau-3B-2, QLr.iau-7A-2, QLr.iau-7A-3 and QLr.iau-7D-2) are more interesting, as they are associated with resistance to two or more Pt races. The known and novel QTLs associated with different Pt races found here, can be used in future wheat breeding programs to recombine different loci for durable resistance against leaf rust races.
Subject terms: Genetics, Plant sciences
Introduction
Bread wheat (Triticum aestivum L.) has been domesticated in Fertile Crescent 10,000 years ago1 and is the most important crop in Iran which is widely cultivated in an area of more than six million hectares. It is suggested that both wheat and its pathogens have co-evolved in this area. Leaf rust incited by Puccinia triticina Erikss. (Pt) is a macrocyclic foliar disease of wheat and is the most widely distributed worldwide and generally appears in most of the wheat-growing regions of Iran particularly at early and late growing stages2. It is believed that the center of origin of P. triticina is the Fertile Crescent region, where the natural range of the primary and alternative hosts overlaps3. P. triticina can cause significant yield losses over large geographical areas and, thus, is considered as a threat of wheat production worldwide4. In Iran, it is the second economic important disease on wheat after yellow rust and under conductive epidemic conditions it is estimated that more than 20% of wheat-growing fields are prone to leaf rust infection5,6.
Different control strategies are currently available to control Pt, including fungicide application, biological control and employment of resistant genes/cultivars. Timely and accurate application of fungicides is effective in controlling of leaf rust in wheat7, but besides the cost of application, fungicides are serious threats to human health and the environment. On the other hand, the reputed use of fungicides may lead to fungicide-resistance in Pt isolates circumventing susceptibility to fungicides7,8. Therefore, characterization of resistance genes and development of resistant cultivars are the most economical and environmentally safe approaches for controlling leaf rust.
P. triticina populations are highly diverse in terms of genetics and virulence pattern, which is driven by the co-evolution of Pt strains with various wheat cultivars in wheat-growing areas worldwide, as well as by genetic recombination of Pt races and spontaneous mutations9,10. High variability in P. triticina populations and its high fitness to diverse environmental conditions results in the regular breakdown of the resistance genes and, hence, the implementation of slow rusting along with race-specific resistance genes has been suggested to enhance the durability of resistance in wheat cultivars4.
Clearly, successful control of leaf rust disease requires basic knowledge about the diversity and virulence profiles of the pathogen populations gained through race analysis approach is necessary for effective control of leaf rust disease. This is critical for establishing effective breeding programs for durable resistance11 . Genetic resistance against leaf rust in wheat is usually related to seedling resistance (referred to all-stage resistance = ASR) and adult-plant resistance (APR)7,12. Seedling resistance is qualitative and controlled by single or major genes that mostly are race-specific resistance and associated with hypersensitive response13,14. While APR is mostly non-race specific resistance and controlled by several minor effect genes, the accuracy of phenotyping for leaf rust under field conditions can be affected by environmental factors such as temperature, light, inoculum pressure and plant maturity12,15.
To date, more than 80 leaf rust resistance genes and QTLs have been identified, of which some were introgressed from durum or bread wheat cultivars and some were originated from wheat wild relatives such as Aegilops, Agropyron, Secale, and Thiropyrum7. So far, a large number of resistance genes and QTLs have been identified in various wheat genotypes16,17. Most of these wheat cultivars and breeding lines, however, are no longer in use because their resistance has been overcome by new virulent Pt races. Therefore, identification of new sources of resistance using different Pt races and implementation of these resistant genotypes into breeding programs are essentially required to control the leaf rust disease18–20.
The objectives of the present study were: (i) to determine the distribution of Pt races in different wheat growing zones of Iran and to monitor the dynamics and variation of virulence to leaf rust resistance genes, (ii) to characterize the resistance/susceptibility pattern in a worldwide collection of wheat genotypes to 10 different Iranian Pt races at seedling stage, and (iii) to conduct genome-wide association analysis (GWAS) for identifying molecular markers associated with known Lr resistance genes and novel QTLs.
Results
P. triticina isolates virulence and race identification
Results of phenotypic interaction of 33 single uredinia of P. triticina isolates on 55 ‘Thatcher’ near-isogenic lines at the seedling stage presented in Table S1. In total 18 physiological races were identified (Table S1). Among all races, FKTPS (15%) and FKTTS (12%) were the most common pathotypes, which were collected mainly from Khuzestan province (southwest of Iran). Phenotypes LKTTS, PJTSS and PKRQS had an occurrence frequency of 9% each. For each DTRRS and PKRQS phenotypes, two isolates were found, while other phenotypes including BKGSS, BRTRS, CFNPs, CFTTS, CTTPR, MFHPs, MHRRS, MJTTS, MTTTS, PTMQS and FSRRS were represented by single isolates.
Geographic distribution of the leaf rust samples is shown in Table S1. The results showed that similar races like DTRRS, FKTTS, LKTTS, PJTSS and PKRQS were isolated from either a single field or geographically close fields. In contrast, FKTPS phenotypes were found from different long-distance locations. Some fields like (Shavoor) contain several races.
Frequencies of virulence to Lr genes or gene combinations were compared. Virulence to Lr2a was not found in any studied area while all isolates were virulent on differentials possessing Lrb, Lr3bg and Lr14b. Virulence to Lr28 was detected only in Kalardasht. Similarly, virulence to Lr9 was found with a low frequency (21%) and was detected in different geographical locations. Virulence to Lr1 was Moderate (48%) while the weighted average frequency of virulence to Lr16, Lr24, Lr3ka, Lr11 and Lr20 was high frequency (> 90%).
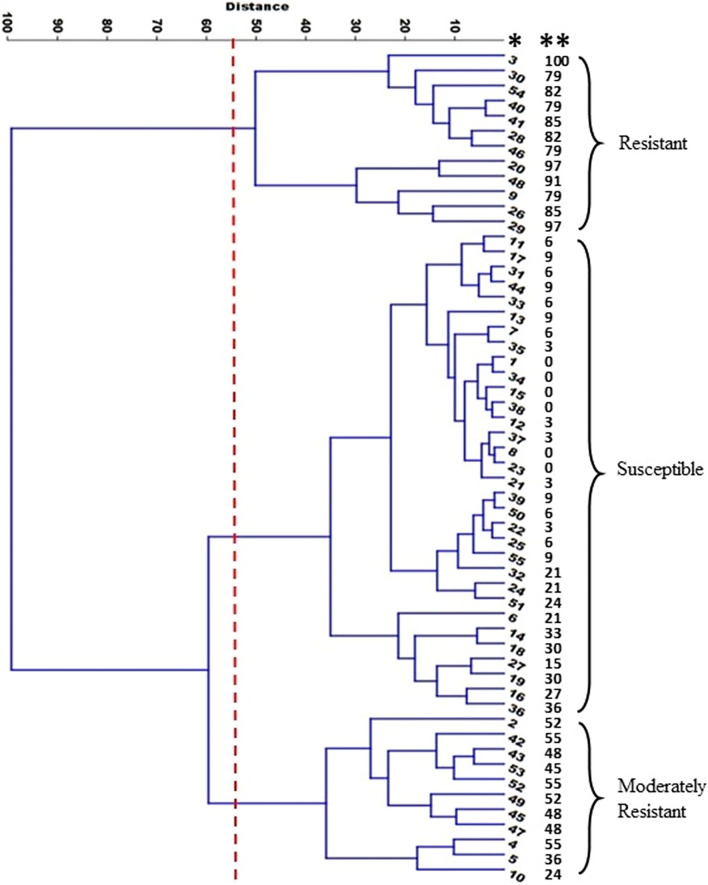
Cluster analysis of wheat differentials showed that wheat genotypes can be categorized into three major clusters (Fig. 1). Cluster A consists of 12 wheat genotypes that were considered resistant. Wheat differentials in this cluster had a high frequency of resistance responses with an average of 86% ranging from 79% (Lr29, Lr23 +, Lr13/Lr17/Lr27 +/Lr31, and Lr9) to 100% (Lr2a) (Fig. 1). Interestingly, in cluster A, Lr2a was resistant to all the tested isolates, which indicates that Lr2a is an effective broad resistance gene. Cluster B is consist of 32 wheat genotypes and was identified as a susceptible cluster with a low average resistance response (11%). Six genotypes in this cluster had no specific resistance responses and were susceptible to all the tested isolates. Resistance responses among all cluster members were generally low ranging from 0 to 36%. Lastly, cluster C showed intermediate responses and was considered as a moderately resistant cluster with an average virulence phenotype of 47% ranging from 24 to 55%.
Figure 1.
Cluster analysis of 55 wheat differential responses against 33 leaf rust isolates showing that wheat genotypes can be categorized into three major clusters including resistant, susceptible and moderately resistant groups. * genotype number, ** Percentage of avirulent isolates.
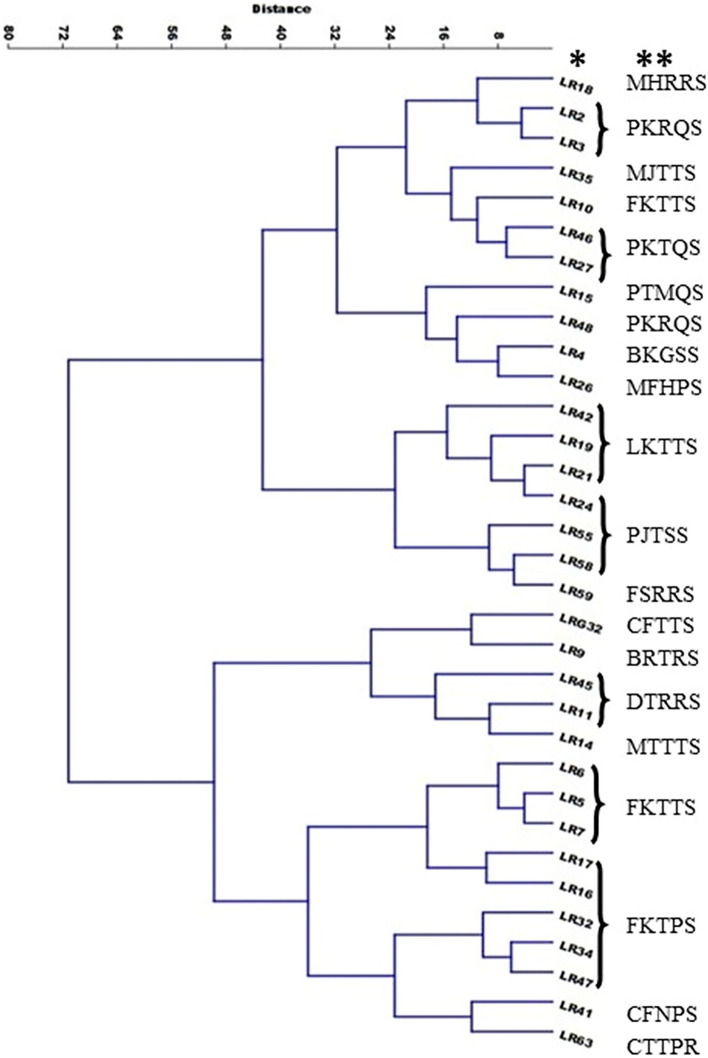
Cluster analysis of 33 leaf rust isolates according to their virulence spectrum on 55 wheat differentials resulted in two main clusters, and each cluster was divided into two sub-clusters. As expected, similar physiologic races grouped together or into very close sub-clusters (Fig. 2). The only exception were isolates from race FKTTS that were separated in two different locations.
Figure 2.
Cluster analysis of 33 isolates according to their virulence spectrum on 55 wheat differentials resulted in several clusters and sub-clusters. Note that the majority of isolates belonging to similar races are grouped in distinct clusters or very close sub-clusters. * Leaf Rust isolates, ** Identified physiologic races.
Wheat germplasm seedling responses to ten Pt races
Infection type (IT) of 185 wheat genotypes against ten Pt races at seedling stage under greenhouse conditions have been presented in Table S2. On the linear scale of 0–9, IT scores ranged from 1 (resistant) to 9 (susceptible), while none of the genotypes showed complete immune responses (IT = 0). Of the ten Pt races, based on IT scores (0–9), less than10% of tested genotypes were found resistant (IT score < 5) to each race, except for FKTPS-1 and FSRRS that 34 (18.3%) and 36 (19.5%) genotypes were resistant, respectively (Table S2). The majority of wheat genotypes used for GWAS analysis showed a high frequency of susceptibility to all races. Based on IT scores, nine genotypes including Oasis (USA), Mehregan (Iran), 40,499 (Australia) and six Iranian advanced breeding lines (ER-M-93-13, ER-N-94-15, ER-S-93-2, ER-S-92-113 and ER-M-92-20) were resistance to all Pt races. Although, an Iranian wheat cv. ‘Parsi’ was resistant to all Pt races except race MTTTS and two landraces (IPK40744 and IPK44673) from USA and India were resistant to all races except races MTTTS and PKRQS (Table S2). Heritability values based on IT scores were high for all Pt races, which means that there was a limited replication variation for phenotypic assessment in relative to genotypic variation. Highly significant positive correlations were observed between Pt races ranging from 0.30 to 0.82, with an average value of 0.56 (Table 1).
Table 1.
Correlation analysis among the phenotypic data of 185 wheat genotypes evaluated for their reaction to 10 Pucnina triticina races.
| Variables | MTTTS | PKRQS | PJTSS | MFHPS | MJTTS | FKTPS‐1 | CTTPR | FKTPS‐2 | FSRRS | BRTRS | Heretability (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MTTTS | 1 | 94.12 | |||||||||
| PKRQS | 0.67** | 1 | 93.8 | ||||||||
| PJTSS | 0.56** | 0.69** | 1 | 91.17 | |||||||
| MFHPS | 0.58** | 0.66** | 0.73** | 1 | 95.14 | ||||||
| MJTTS | 0.30* | 0.41** | 0.40* | 0.36* | 1 | 93.18 | |||||
| FKTPS-1 | 0.44* | 0.47** | 0.38* | 0.36* | 0.74** | 1 | 92.16 | ||||
| CTTPR | 0.50** | 0.55** | 0.53** | 0.59** | 0.46* | 0.55** | 1 | 92.87 | |||
| FKTPS-2 | 0.49** | 0.59** | 0.68** | 0.82** | 0.33* | 0.35* | 0.58** | 1 | 94.16 | ||
| FSRRS | 0.53** | 0.64** | 0.69** | 0.78** | 0.42* | 0.39* | 0.57** | 0.82** | 1 | 93.58 | |
| BRTRS | 0.59** | 0.66** | 0.72** | 0.73** | 0.47* | 0.45* | 0.61** | 0.74** | 0.79** | 1 | 93.74 |
* Significant (α = 0.05); ** Significant (α = 0.01)
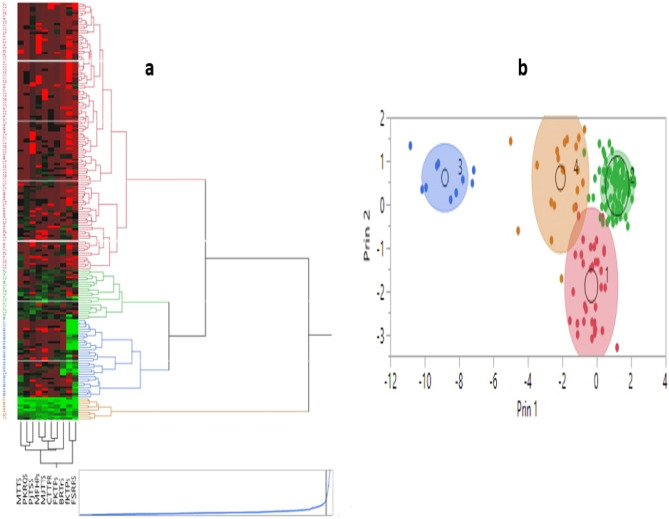
Cluster analysis and principal component analysis based on IT scores (0–9) grouped 185 wheat genotypes into four clusters (Fig. 3). The first cluster comprised 118 wheat genotypes, which showed high susceptibility to all Pt races (Table 2). Cluster-II contained 22 genotypes, most of which were landraces from different sources and also included four Iranian improved cultivars. None of this cluster genotypes showed high resistance to all races, while a few of them only showed resistant spectra to FKTPS-1, PJTTS or FSRRS races (Table S2). Cluster III comprised 35 genotypes, all of which were Iranian improved cultivars and advanced breeding lines. These genotypes showed resistance spectra to FKTPS-1 and FSRRS races (Table 2). Cluster IV contained 10 genotypes, including six Iranian advanced breeding lines, one Iranian Cultivar ‘Mehregan’, a cultivar originating from the USA ‘Oasis’ and two landraces IPK40499 and IPK44673 from Australia and India, respectively. These genotypes showed a high level of resistance to all Pt races (Table 2; Table S2).
Figure 3.
Cluster analysis (a) and principal component analysis (PCA) (b) of 185 wheat genotypes based on IT scores data against ten Pucnina. triticina races.
Table 2.
Means of disease severity (0–9) of wheat genotypes to different P.triticina races in four clusters.
| Cluster | No. of genotypes | MTTS | PKRQS | PJTSS | MFHPS | FKTPS‐1 | FSRRS | BRTRS | MJTTS | CTTPR | FKTPS-2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 118 | 7.67 | 7.53 | 7.97 | 7.88 | 8.08 | 7.54 | 7.57 | 7.82 | 7.75 | 7.87 |
| 2 | 22 | 6.59 | 5.95 | 6.64 | 5.77 | 6.27 | 5.68 | 6.18 | 5.32 | 5.95 | 6.27 |
| 3 | 35 | 6.66 | 6.86 | 7.94 | 7.91 | 3.77 | 2.54 | 6.17 | 7.97 | 7.66 | 7.43 |
| 4 | 10 | 3.10 | 2.90 | 3.00 | 2.30 | 1.90 | 1.60 | 2.20 | 1.90 | 2.20 | 2.10 |
Wheat panel diversity, population structure and LD analysis
Genotyping of 185 wheat genotypes returned a total of 94,535 raw DArT-seq markers. After marker filtering, in total 21,773 DArTseq markers (including 15,856 SilicoDArT and 5917 SNP) with MAF ≥ 5% and missing data points ≤ 20%, were used for further analysis of population structure, linkage disequilibrium and marker-trait association analysis against 10 Pt races. Analysis of genetic diversity using UNJ-clustering and Bayesian model-based structure of 185 wheat genotypes used in this study was previously described by Maboubi et al. (2022), where this wheat panel was grouped into four distinct clusters (Fig. S1). This cluster grouping was relatively consistent with the geographical origin and type (landrace or cultivar) of genotypes. A comparable result similar to population structure and UNJ-clustering was also observed by the heatmap plot of the kinship matrix where four distinct clusters were identified (Fig. S2). The first cluster comprised 50 genotypes, of which 17 were Iranian landraces and as well as landraces originating from Turkey, Romania, Hungary and Tajikistan. All of these genotypes were susceptible to most of the Pt races, except a few Iranian landraces that showed partial resistance to FKTPS-1 race. Cluster-II comprised of 29 genotypes from globally diverse origins and were susceptible to all Pt races. Cluster-III included 86 genotypes, which most of the Iranian cultivars grouped in this cluster. Most of the genotypes in this cluster, with a few exceptions, were resistant to two Pt races (FKTPS-1 and FSRRS). Cluster-IV comprised of 20 genotypes, of which were mostly resistant to multiple Pt races and ten wheat genotypes that showed a high resistance pattern to all Pt races were in this cluster. The results of population genetic diversity were significantly in agreement with phenotypic responses of wheat genotypes against Pt races, with a few exceptions, which indicated the good fit of population structure analysis with phenotypic data that are prerequisite for marker-trait association analysis. In the LD analysis, 28% of the intra-chromosomal pairs showed a significant level (P < 0.001) of the correlation coefficient (r2). Mean and critical r2 values were 0.09 and 0.16, respectively. Overall, LD between marker pairs decayed quickly in the B genome, followed by the A genome. In the D genome the LD was very pronounced and they did not drop below the critical value over distances of 1.6 kb (Fig. S3).
Association mapping against Pt races and putative candidate gene identification
The GWAS based on normalized IT scores to ten Pt race/isolate at seedling stages using Farm-CPU model showed reliable results and presented low spurious associations. Association analysis was performed separately for each Pt race. A total of 62 significant markers were identified and found distributed across all chromosomes except for 1D, 3A, 4B, 4D and 5D (Table 3 and Fig.S4). The significant markers explained 6–18% of phenotypic variations. The QTLs identified for different Pt races but located at an overlapping genomic region on a chromosome were considered a single QTL and assigned the same name using the nomenclature QLr.iau- followed by the name and number of QTL in chromosome order and finally, 62 MTAs were assigned to 34 QTL regions on 16 chromosomes (Table 3).
Table 3.
Summary of the seedling leaf rust resistance quantitative trait loci identified against 10 Puccinia triticina races in the panel of 185 wheat genotypes.
| QTL | SNP ID | RACE | Chr | Position (bp) | Position (cM) | P value | Annotated gene | Predicted function |
|---|---|---|---|---|---|---|---|---|
| QLr.iau‐1A‐1 | 1,104,046 | MJTTS | chr1A | 31,875,119 | 45.11 | 4.11E − 05 | TraesCS1A02G051100 | Leucine-rich repeat domain superfamily |
| QLr.iau‐1A‐2 | 1,091,963 | MFHPS | chr1A | 544,082,192 | 153.82 | 6.84E − 05 | TraesCS1A02G366500 | Cytochrome P450 |
| QLr.iau‐1A‐3 | 3,022,780 | MJTTS | chr1A | 588,634,744 | 252.78 | 5.65E − 05 | TraesCS1A02G440300 | Leucine-rich repeat-containing N-terminal |
| QLr.iau‐1B‐1 | 1,104,236 | MJTTS | chr1B | 479,758,475 | 94.37 | 1.78E − 06 | TraesCS1B02G274400 | Protein kinase-like domain (Zinc finger, RING/FYVE/PHD-type) |
| QLr.iau‐1B‐2 | 1,696,203 | MJTTS | chr1B | 682,864,419 | 276.30 | 7.88E − 05 | TraesCS1B02G474600 | F-box-like domain superfamily |
| QLr.iau‐2A‐1 | 3,938,806 | FSRRS | chr2A | 6,622,475 | 9.45 | 2.61E − 06 | TraesCS2A02G016900 | P-loop nucleoside triphosphate hydrolase |
| QLr.iau‐2A‐2 | 3,027,084 | MTTTS | chr2A | 24,111,039 | 65.85 | 2.25E − 05 | TraesCS2A02G057000 | Protein kinase-like superfamily (Legume lectin domain) |
| 4,261,248 | PKRQS | chr2A | 182,140,338 | 67.79 | 1.20E − 06 | TraesCS2A02G204700 | P-loop containing nucleoside triphosphate hydrolase (Plant myosin class VIII) | |
| 3,021,874 | FKTPS‐2 | chr2A | 389,439,783 | 68.56 | 3.22E − 05 | TraesCS2A02G255300 | Leucine-rich repeat domain superfamily | |
| QLr.iau‐2A‐3 | 981,785 | MFHPS | chr2A | 742,781,080 | 110.47 | 4.32E − 05 | TraesCS2A02G520500 | Cytochrome P450 superfamily |
| 3,024,004 | CTTPR | chr2A | 769,344,169 | 122.19 | 1.39E − 06 | TraesCS2A02G573500 | Haem peroxidase superfamily | |
| QLr.iau‐2B‐1 | 995,662 | MTTTS | chr2B | 18,319,883 | 13.54 | 7.24E − 05 | TraesCS2B02G038400 | Leucine-rich repeat-containing N-terminal, plant-type |
| 3,532,895 | MTTTS | chr2B | 28,176,051 | 24.84 | 3.10E − 05 | TraesCS2B02G058900 | Leucine-rich repeat domain (Virus X resistance protein-like) | |
| 3,020,982 | PJTSS | chr2B | 48,007,814 | 32.82 | 4.31E − 05 | TraesCS2B02G085400 | Leucine-rich repeat domain superfamily | |
| QLr.iau‐2B‐2 | 1,027,810 | MFHPS | chr2B | 551,637,948 | 76.50 | 9.73E − 05 | TraesCS2B02G388800 | (Legume lectin domain) |
| 1,862,545 | PKRQS | chr2B | 623,869,418 | 77.50 | 4.88E − 05 | TraesCS2B02G430900 | Plant Peptidase S10 | |
| 3,946,214 | FKTPS‐2, | chr2B | 642,834,436 | 78.31 | 4.10E − 05 | TraesCS2B02G450200 | Cytochrome P450 | |
| QLr.iau‐2B‐3 | 1,152,655 | CTTPR, MFHPS, MJTTS | chr2B | 712,050,374 | 128.14 | 2.38E − 06 | TraesCS2B02G517300 | Leucine-rich repeat domain superfamily |
| QLr.iau‐2D‐1 | 3,940,894 | MTTTS | chr2D | 5,283,967 | 6.06 | 8.29E − 05 | TraesCS2D02G012800 | Cytochrome P450 |
| QLr.iau‐3B‐1 | 1,088,335 | CTTPR | chr3B | 518,350,866 | 60.83 | 7.43E − 09 | TraesCS3B02G320700 | Leucine-rich repeat domain superfamily |
| 1,231,107 | CTTPR | chr3B | 653,810,881 | 73.15 | 4.09E − 05 | TraesCS3B02G418800 | F-box-like domain superfamily | |
| 5,369,257 | FKTPS‐2 | chr3B | 671,161,431 | 78.81 | 1.79E − 05 | TraesCS3B02G433100 | Leucine-rich repeat domain superfamily | |
| 1,216,374 | FKTPS‐2 | chr3B | 671,161,607 | 78.81 | 7.11E − 05 | – | – | |
| QLr.iau‐3B‐2 | 1,229,647 | PJTSS | chr3B | 797,786,785 | 131.01 | 4.27E-05 | TraesCS3B02G565900 | Leucine-rich repeat domain superfamily |
| 1,076,425 | BRTRS | chr3B | 812,658,362 | 138.38 | 5.67E − 05 | TraesCS3B02G586500 | Cytochrome P450 | |
| 1,057,473 | MFHPS | chr3B | 812,993,842 | 145.09 | 5.83E − 05 | – | – | |
| 1,095,941 | MFHPS | chr3B | 813,391,786 | 145.09 | 4.35E − 05 | TraesCS3B02G587400 | Leucine-rich repeat domain superfamily | |
| 4,989,676 | MFHPS | chr3B | 824,481,249 | 156.69 | 3.29E − 06 | TraesCS3B02G606700 | Leucine-rich repeat domain superfamily | |
| 1,111,693 | CTTPR | chr3B | 827,961,228 | 156.74 | 9.02E − 05 | TraesCS3B02G609300 | F-box-like domain superfamily | |
| QLr.iau‐3D‐1 | 1,003,778 | PKRQS | chr3D | 544,419,623 | 89.14 | 1.17E − 05 | TraesCS3D02G430100 | Leucine-rich repeat domain superfamily |
| QLr.iau‐4A‐1 | 3,936,450 | BRTRS | chr4A | 153,654,485 | 26.46 | 1.27E − 06 | TraesCS4A02G123700 | P-loop nucleoside triphosphate hydrolase |
| 5,967,805 | MFHPS | chr4A | 202,953,887 | 27.42 | 6.14E − 05 | – | – | |
| QLr.iau‐4A‐2 | 995,761 | MFHPS, PJTSS,PKRQS | chr4A | 607,270,056 | 54.26 | 7.25E − 06 | TraesCS4A02G318300 | Leucine-rich repeat domain superfamily |
| QLr.iau‐4A‐3 | 1,351,280 | FKTPS‐2 | chr4A | 629,433,955 | 88.61 | 3.96E − 05 | TraesCS4A02G355800 | Cytochrome P450 |
| QLr.iau‐4A‐4 | 1,233,446 | CTTPR | chr4A | 708,659,775 | 116.03 | 1.40E − 06 | TraesCS4A02G438700 | Leucine-rich repeat domain superfamily |
| QLr.iau‐5A‐1 | 1,703,104 | PJTSS | chr5A | 436,890,364 | 42.45 | 6.60E − 06 | TraesCS5A02G222200 | Leucine-rich repeat domain superfamily |
| 2,277,102 | MJTTS | chr5A | 503,499,615 | 54.79 | 1.14E − 05 | TraesCS5A02G294800 | Protein kinase-like domain superfamily | |
| QLr.iau‐5B‐1 | 1,067,819 | PJTSS | chr5B | 6,391,001 | 0.00 | 8.27E − 05 | TraesCS5B02G004500 | Cytochrome P450 |
| QLr.iau‐5B‐2 | 4,261,927 | CTTPR | chr5B | 330,119,186 | 29.60 | 1.97E − 06 | TraesCS5B02G181000 | F-box-like domain superfamily |
| QLr.iau‐5B‐3 | 4,911,101 | MFHPS | chr5B | 528,472,570 | 54.18 | 1.93E − 05 | TraesCS5B02G341300 | F-box-like domain superfamily |
| QLr.iau‐5B‐4 | 1,067,151 | PJTSS | chr5B | 704,868,810 | 139.40 | 4.04E − 07 | TraesCS5B02G554300 | Leucine-rich repeat domain superfamily |
| 2,258,090 | PKRQS | chr5B | 706,684,737 | 146.28 | 4.80E − 06 | TraesCS5B02G560200 | Leucine-rich repeat domain superfamily | |
| QLr.iau‐6A‐1 | 3,064,900 | MJTTS | chr6A | 39,002,398 | 33.61 | 4.83E − 05 | TraesCS6A02G071900 | Serine-threonine/tyrosine-protein kinase |
| 1,045,339 | BRTRS | chr6A | 371,813,592 | 48.63 | 6.22E − 05 | – | – | |
| QLr.iau‐6B‐1 | 2,276,989 | MFHPS | chr6B | 366,585,056 | 30.36 | 6.20E − 05 | – | – |
| 985,117 | FKTPS | chr6B | 485,290,761 | 30.95 | 2.12E − 05 | TraesCS6B02G269500 | Cytochrome P450 superfamily | |
| 1,003,530 | CTTPR | chr6B | 581,549,680 | 33.12 | 1.56E − 06 | – | – | |
| 996,529 | PJTSS | chr6B | 669,396,201 | 49.88 | 5.67E − 05 | TraesCS6B02G394600 | Leucine-rich repeat domain superfamily | |
| QLr.iau‐6D‐1 | 992,973 | PJTSS | chr6D | 5,175,026 | 7.39 | 3.28E − 05 | TraesCS6D02G012900 | Leucine-rich repeat domain superfamily |
| QLr.iau‐7A‐1 | 3,021,075 | CTTPR | chr7A | 39,176,390 | 28.66 | 1.81E − 08 | TraesCS7A02G074600 | hydrolase (Helicase superfamily) |
| 1,102,645 | CTTPR | chr7A | 107,080,713 | 39.64 | 1.21E − 05 | TraesCS7A02G155000 | Cytochrome P450 superfamily | |
| QLr.iau‐7A‐2 | 5,373,057 | FKTPS‐2, PJTSS | chr7A | 301,167,038 | 88.73 | 4.79E − 05 | – | – |
| 1,094,354 | MTTTS | chr7A | 651,881,285 | 97.58 | 1.60E − 05 | TraesCS7A02G455800 | Protein kinase-like domain superfamily (Serine/threonine-protein kinase) | |
| QLr.iau‐7A‐3 | 1,103,172 | MJTTS | chr7A | 705,917,325 | 149.56 | 4.15E − 05 | TraesCS7A02G522300 | Cytochrome P450 superfamily |
| 1,109,797 | BRTRS | chr7A | 724,430,683 | 153.97 | 7.58E − 05 | TraesCS7A02G550000 | Leucine-rich repeat domain superfamily | |
| QLr.iau‐7B‐1 | 2,275,239 | CTTPR | chr7B | 210,779,344 | 42.30 | 7.18E − 08 | TraesCS7B02G157000 | P-loop nucleoside triphosphate hydrolase (Phosphoribosyltransferase-like) |
| 1,019,331 | BRTRS, FSRRS | chr7B | 220,270,676 | 45.11 | 4.85E − 05 | TraesCS7B02G162500 | Protein kinase-like domain superfamily | |
| 1,066,279 | FKTPS | chr7B | 263,698,243 | - | 3.85E − 05 | TraesCS7B02G179000 | Papain-like cysteine peptidase superfamily | |
| QLr.iau‐7B‐2 | 1,017,404 | FKTPS | chr7B | 688,713,574 | 99.89 | 1.67E − 05 | TraesCS7B02G419600 | Leucine-rich repeat domain superfamily |
| QLr.iau‐7D‐1 | 4,910,573 | FKTPS‐2 | chr7D | 41,889,844 | 41.31 | 1.32E − 05 | TraesCS7D02G072300 | P-loop nucleoside triphosphate hydrolase (Kinesin-like protein) |
| QLr.iau‐7D‐2 | 1,079,705 | FKTPS‐2 | chr7D | 420,047,431 | 100.64 | 4.01E − 06 | TraesCS7D02G331500 | Leucine-rich repeat domain superfamily |
| 3,959,264 | MTTTS | chr7D | 458,680,743 | 104.63 | 9.28E − 05 | TraesCS7D02G354700 | Leucine-rich repeat-containing N-terminal, plant-type |
Most of significant regions (QTLs) were associated with resistance to multiple races, although 18 QTLs showed race-specific resistance on chromosome 1A (QLr.iau-1A-1, QLr.iau-1A-2 and QLr.iau-1A-3), IB (QLr.iau-1B-1 and QLr.iau-1B-2), 2A (QLr.iau-2A-1), 2B (QLr.iau-2B-3), 2D (QLr.iau-2D-1), 3D (QLr.iau-3D-1), 4A (QLr.iau-4A-2, QLr.iau-4A-3 and QLr.iau-4A-4), 5B (QLr.iau-5B-1, QLr.iau-5B-2 and QLr.iau-5B-3), 6D (QLr.iau-6D-1), 7B (QLr.iau-7B-2) and 7D (QLr.iau-7D-1) (Table 3). The large effect loci on chromosomes 2A, 2B, 3B, 4A, 5B and 7A were associated with responses to multiple Pt races. Resistance-associated QTLs localized on 8 and 7 different chromosomes were identified for CTTPR and MFHPS races, respectively, while for FSRRS/FKTPS and BRTRS races, resistance-associated QTLs were identified only on two and three chromosomes, respectively (Table 3). In this study, two isolates belonging to FKTPS race were used. For FKTPS-2 (originated from Ahvaz, southwest of Iran) multiple QTLs localized on six different chromosomes were identified. Interestingly, all QTLs against FKTPS-1 (originated from Neishaboor, north-east of Iran) were different from those QTLs identified for FKTPS-2.
The chromosomal position of MTAs associated with resistance to Pt races were mapped to the Chinese Spring cv. wheat physical genome. For each MTA, 2.5 Mb region toward the left and right side was used to identify the putative candidate genes. Totally, in 56 MTAs we identified several putative candidate genes previously known to play a role in defense mechanisms such as genes encoding leucine-rich repeat (LRR), protein kinase, zinc finger and P-loop-NTPase proteins (Table 3).
Discussion
Pathotypes and physiologic specialization of Iranian wheat leaf rust
Wheat is the most important cereal food crop worldwide and it has been domesticated and cultivated in Iran from ancient times2. Leaf rust caused by P. triticina Eriks (Pt), is the most important and common foliar disease of wheat in Iran and most wheat growing area worldwide3,21. Given the fact that both wheat and P. triticina have coevolved in Near-East as well as in Iran, so this fungi has probably been present in this area for thousands of years3.
In this study, 33 P. triticina isolates from different wheat-growing areas of Iran were collected and tested for race determination based on their reaction on 55 differential wheat genotypes possessing different Lr resistance genes. Our results showed that most of the resistance genes were ineffective against P. triticina population. However, low virulence phenotype on Lr2a, Lr9, Lr19, Lr25, Lr28, and Lr29 indicates that these genes are still effective against the wheat leaf rust population in Iran at present, which is basically consistent with previous studies3,22,23. For example, leaf rust surveys conducted in Iran from 2002 to 2004 indicated no virulence for Lr9, Lr18, Lr19, Lr25, Lr28, Lr29, Lr34, Lr35, Lr36, or Lr37 in the field22. In addition, no virulence to Lr2a, Lr3ka, Lr9, Lr14a, Lr19, Lr23, Lr25, Lr26, Lr28, Lr29, Lr30, Lr32, or Lr36 was detected in 2008. Furthermore, race analysis in 2009 and 2010 showed that virulence to Lr9, Lr28, Lr25, Lr19, Lr29, and Lr2a were at low frequencies23.
It is believed that forces of mutation, migration, sexual and asexual recombination and selection pressure play significant roles in pathogenic diversity and appearance of new races of rust diseases24. Like other rust diseases, urediniospores of leaf rust could migrate thousands of kilometers causing exotic races and clonal reproduction3,25.
Recent studies report the similarity between some Iranian and Russian P. triticinia isolates that might be attributed to northerly winds that blow from Russia to the north of Iran2. On the contrary, our results showed large difference between the phenotypes of Iranian and Russian isolates. None of the virulence phenotypes across Russia had virulence on the leaf rust resistance genes Lr24 or Lr28 and phenotypes with virulence on Lr16 and Lr18 were at frequencies < 10% of total isolates and were not present in all regions. Interestingly, unlike Iranian isolates that were avirulent to Lr2a¸virulence phenotypes on this gene was found at high frequency (66%)26. In Pakistan virulence to Lr9, Lr19 or Lr28 was not identified27 that is partly similar to what was observed in this study. However, recently virulence on Lr2a was identified28, which is different from Iranian isolates. In other neighboring countries like Armenia, Azerbaijan, Tajikistan, Kazakhstan, Uzbekistan, and Kyrgyzstan that are located in North of Iran (Central Asia), no virulence on Lr9, Lr23, Lr24 or Lr26 was found29. While no virulence’s were detected against Lr12, Lr15, Lr17, Lr22a and Lr24 in Iraq30. In Syria, no virulence for Lr1, Lr2a, Lr9, Lr15, Lr19, Lr21, Lr24, Lr25, Lr26, Lr28, or Lr29 was observed in greenhouse tests showing that Syrian isolates were less aggressive than those of leaf rust isolates in this region31.
This section of our findings provided detailed information on the variation in virulence patterns of Iranian P. triticina isolates, a country located in the Fertile Crescent where wheat domestication began and coincided with the speciation and further evolution of its pathogens32. We demonstrate that P. triticina isolates have a broad virulence spectrum against most of the known Lr genes indicating that extensive host adaptation has occurred in P. triticina populations during the synchronic domestication process of both host and pathogen in this region. Our data shed light into the potential employment of the effective Lr genes like Lr2a, Lr28 and Lr19 that are of interest to wheat breeding programs to improve the resistance of Iranian wheat cultivars against leaf rust.
Novel resistance sources and alignment with previously reported QTLs and Lr genes
Characterization of novel resistance sources is the prerequisite and most important strategy for controlling rust diseases in wheat and pyramiding these genes for durable resistance33,34. The rapid evolution of pathogens due to fungicide application, environmental conditions and also narrow genetic base of resistance genes in improved wheat genotypes can easily lead to the breakdown of the resistance genes17,35. In this study, 185 wheat genotypes comprising Iranian cultivars and landraces from diverse world geographical origins were evaluated for resistance against 10 Pt races at seedling stages. Based on IT scores, only three cultivars (Oasis, Mehregan and Parsi) and six Iranian advanced breeding lines showed resistance to all Pt races. Interestingly, all of these genotypes originated from Iran, except for Oasis originated from the USA. The resistance frequency of wheat genotypes for most of the P. triticina races was very low (~ 15%), except for two races (FKTPS-1 and FSRRS). Therefore, the resistance pattern of the studied wheat germplasm did not correspond to the virulence profiles of the P. triticina races as we identified on the set of 55 wheat differential genotypes. It can be concluded that these wheat genotypes may carry multiple previously known Lr genes or in combination with new genes17,36.
Significant positive correlations were observed for infection types of ten P. triticina races (Table 1). This can conclude that by pathogenicity test results of these ten races on a set of 55 differential genotypes (Table S1), in which all Pt races were virulent on Lr3, Lr11, Lr12, Lr13, Lr20, Lr21, Lr33, Lr34, Lr35 and Lr37. On the other hand, it is likely that the wheat panel used for the GWAS had multiple genomic loci conferring resistance to multiple races, which was further confirmed by the results of association mapping analysis (Table 3). Similar results for significant phenotypic correlation between multiple races of P. triticina in different GWAS panel have been reported14,37,38. Therefore, to elucidate the genetics of resistance to P. triticina in the wheat panel we implemented a high-throughput genome association analysis using DArTseq markers against 10 Pt races.
Overall GWAS analysis using different races identified 62 MTAs that were assigned to 34 QTL regions on 16 chromosomes (Table 3). these genomic regions were compared with the previously known leaf rust resistance (Lr) genes and QTLs projected on consensus maps39,40 (Fig. S5). Three QTLs on chromosome 1A were race-specific for resistance to MJTTS and MFHPS races. Two QTLs (QLr.iau-1A-1 and QLr.iau-1A-2) co-located with previously known adult plant resistance (APR) QTLs41–43. The QTL Qlr.iau-1A-3 (252.78 cM) was detected for resistance to MJTTS race and did not align with any previously reported QTL or Lr genes, therefore it considered as a potential novel QTL. Two QTLs on chromosome 1B were race-specific for resistance to MJTTS including QLr.iau-1B-1 was co-localized with different previously known APR resistance QTLs44–48 as well as with four known resistance genes Lr33, Lr44, Lr71 and Lr7549. The QTL QLr.iau-1B-2 (276.30 cM) mapped on 1BL chromosome, but its chromosomal location is far (50 cM) from recently reported APR resistance QTL as well as a QTL found against THBL race form the USA on this chromosome arm12,50. Therefore, this region can be considered as a novel locus for race-specific resistance to MJTTS. Both of the QTLs on chromosome 1B were associated with the resistance to MJTTS, but localized on different arms.
Three QTLs were detected on chromosome 2A, of which QLr.iau-2A-1 was race-specific for resistance to FSRRS and the other two QTLs were detected against multiple races. All these QTLs were co-localized with previously known APR resistance QTLs12,44,51,52. Three QTLs were detected on chromosome 2B, of which two QTLs (QLr.iau-2B-1 and QLr.iau-2B-2) were associated with multiple Pt races and co-localized with previously known resistance genes (Lr18 and Lr37) and QTLs53. QLr.iau-2B-3 was race-specific for resistance to MJTTS and co-localized with previously known QTLs at the adult plant stage47,54.
Two genomic loci were detected on chromosome 3B, of which QLr.iau-3B-1 was associated with resistance to two races, CTTPR and FKTPS. This QTL co-localized with previously known QTLs associated with APR resistance in the field46,55,56. Interestingly, another QTL (QLr.iau-3B-2) was associated with resistance to multiple races (PJTSS, BRTRS, MFHPS and CTTPR) and did not align with any previously reported QTL or Lr genes, therefore we assume this QTL might be as a potential novel QTL. The QLr.iau-3D-1 QTL was associated with the race-specific resistance to PKRQS race and did not align with any previously QTLs on this chromosome. Given the fact that no resistance Lr gene except a few QTL for leaf rust resistance identified on chromosome 3D, further studies are needed to elucidate these loci for resistance to more races and also for finding the exact position with more closely significant markers in this region.
Four genomic loci were detected on chromosome 4A, of which two QTLs (QLr.aiu-4A-1 and QLr.aiu-4A-2) were associated with multiple races and co-localized with previously known QTLs and Lr3057,58. In addition, two QTLs, QLr.aiu-4A-3 and QLr.aiu-4A-4 were associated with race-specific resistance to FKTPS and CTTPR, respectively. These genomic loci were not aligned with any previously identified QTLs or Lr genes, therefore, we concluded that these are potential novel QTLs. Four race-specific genomic loci on chromosome 5B were identified to be associated with different races and three QTLs on chromosomes 5A, 6A and 6B were associated with resistance to multiple races. All these QTLs co-localized with previously known QTLs for adult plant resistance42,47,52,55,58–60.
A QTL QLr.iau-6D-1 on chromosome 6D was associated with race-specific resistance to PJTSS and did not align with previous reported QTLs on this chromosome. So far no Lr gene has been identified, and only a few QTLs for leaf rust resistance have been identified on this chromosome45, which did not align with QLr.iau-6D-1 indicating that QLr.iau-6D-1is a potential novel QTL, which needs to be further investigated.
Three QTLs were identified on chromosome 7A, of which QTL QLr.aiu-7A-1 was associated with resistance to CTTPR and FKTPS races and co-localized with previously reported adult plant resistance QTLs reported12 and Lr47, which is a seedling leaf rust resistance gene that introgressed from Triticum speltoides into the bread wheat genome61. Another two QTLs, QLr.iau-7A-2 and QLr.iau-7A-2 were also associated with resistance to multiple races and did not align with previously known QTLs or Lr genes, which can be considered as potential novel QTLs. Two QTLs on chromosome 7B were associated with resistance to multiple races and co-localized with previously known APR resistance QTLs42,62. Two QTLs identified on chromosome 7D, of which QLr.iau-7D-1 co-localized with previously reported leaf rust QTLs63,64. QTL QLr.iau-7D-2 associated with resistance to FKTPS and MTTTS races mapped at a distance of ≥ 20 cM from L34. According to the pathogenicity test of ten P. triticina races used in this study on 55 differential genotypes, all of them were virulent to Lr34. Therefore, QLr.iau-7D-2 is unlikely to be Lr34, which can be considered as a novel QTL.
Conclusions
High numbers of P. triticina races detected in this study from different wheat growing areas in Iran showed a relatively high diversity of Pt isolates/races that could be due to migrations of this pathogen from neighboring countries like Russia, Turkey and Iraq to Iran. Different virulence patterns of these isolates against wheat differentials indicated that some Lr genes like Lr2a, Lr9, Lr25, Lr28 and Lr29 are still effective against Iranian Pt races and can be used in breeding programs. Results of GWAS analysis on 185 worldwide wheat genotypes using 10 Pt races, identified 34 QTLs, of which 18 were race-specific and 14 QTLs were associated with resistance to two or more P. triticina races. Consequently, 10 loci on chromosomes 1A, 1B, 3B, 3D, 4A, 6D, 7A and 7D were identified as potential novel QTLs. Four of those (QLr.iau-3B-2, QLr.iau-7A-2, QLr.iau-7A-3 and QLr.iau-7D-2) are more interesting, as they are associated with resistance to two or more Pt races. Most of the identified QTLs in this study were co-localized with previously known APR resistance QTLs. Our finding can be used for combining seedling resistance with APR QTLs/genes, which is an effective and promising strategy for durable leaf rust resistance in wheat.
Materials and methods
P. triticina isolation and propagation
During the spring and summer of 2016, naturally infected wheat fields from 10 provinces and 18 distant wheat-growing locations were surveyed. In general from each location 2–4 leaf rust samples were collected, air-dried, and temporarily stored at 4 °C in a refrigerator until later use (Table S1). From each sample a single P. triticina uredinia was isolated, purified and used for further investigation. To do so, the dried leaves were placed on wet filter papers in Petri dishes and kept at 20 °C overnight. Uredinia were then inoculated onto 10-day-old seedlings of Iranian susceptible cv. Boolani. After inoculation, wheat plants were then transferred to a dark room overnight at %100 relative humidity (25 °C) and then were returned and maintained in a greenhouse at 20–25 °C with supplemental fluorescent lighting to provide a photoperiod of 16 h with a light density of 16,000 lx. Single pustules were derived from each sample after 14 days post inoculation and were increased on susceptible seedling plants again using the same procedure. Isolates were collected by vacuum collectors or by tapping wheat leaves having uredinia. Uredinia were dried in a desiccator containing silica gel for two days and stored at − 80 °C for later use.
Race and virulence identification
The first experiment for P. triticina race and virulence identification was carried out at the Cereal Research Department, Seed and Plant Improvement Institute (SPII), Karaj, Alborz, Iran, in 2018. This experiment included 33 P. triticina isolates, which were tested on 55 near-isogenic Thatcher wheat lines (each comprising a single resistance gene). These included set 1: Lr1, Lr2a, Lr2c and Lr3, set 2: Lr9, Lr16, Lr24 and Lr26, set 3: Lr3ka, Lr11, Lr17 and Lr3065, set 4: Lrb, Lr10, Lr14a and Lr1866 and set 5: Lr3bg, Lr14b, Lr20 and Lr2813. In addition, a set of other resistant lines each possessing multiple Lr genes in the different genetic background was used (Table S1). To conduct virulence assay, three pots (as a three replications) of each genotype contained 6–10 seeds in each pots were planted and 10-days old seedling plants were inoculated with uredinia of each isolate suspended in mineral oil (0.3 ml L−1) at the concentration of 6 × 105 spores/ml67.
The infection types (IT) on the primary leaves were recorded at 14 days post-inoculation, when uredinia on susceptible cultivar were fully developed using 0-to-4 scaling system as described previously24,68. Infection types 0 to 2 + were considered to show avirulence for a particular Lr gene and infection types 3 to 4 virulence. Based on the low or high infection types of each isolate on the 55 wheat near-isogenic Thatcher lines, a five-letter code for each race was designated using the North American letter code nomenclature system69. Cluster analysis of IT data for both wheat differentials and isolates was done based on the dissimilarity matrix calculated with the Manhattan index, as implemented in the PAST software v.1.9370.
Leaf rust seedling response assays in 185 wheat genotypes
In the second experiment, phenotyping evaluation of an AM panel consisting of 185 worldwide diverse wheat genotypes (Table S2) was carried out at the Cereal Research Department, Seed and Plant Improvement Institute (SPII), Karaj, Iran, in 2018–2019. Wheat genotypes were tested at the seedling stage under greenhouse conditions using a randomized complete block design with two replications against 10 Pt races. The Pt races were chosen according to different virulence patterns (Table 4) of isolates from distinct geographical regions based on the results of the first experiment. Experimental procedures for inoculation, incubation and disease assessment were the same as those described for race identification in near-isogenic Thatcher lines, using a 0-to-4 scaling system as described previously24,68.
Table 4.
Physiological race and collection site of 10 Pucnina. triticina isolates used for phenotypic assessment at the seedling stage on 185 wheat genotypes.
| Race | Isolate | Origin (Province/City) |
|---|---|---|
| MTTTS | LR15 | Khuzestan/Shavoor |
| PKRQS | LR2 | Khuzestan/Dezful |
| PJTSS | LR59 | Ardabil/Ardabil |
| MFHPS | LR42 | Khorasan Razavi/Mashhad |
| MJTTS | LR35 | Mazandaran/Behshahr |
| FKTPS-1 | LR47 | Khorasan Razavi/Neishaboor |
| CTTPR | LR63 | Mazandaran/Kalardasht |
| FKTPS-2 | LR16 | Khuzestan/Ahvaz |
| FSRRS | LRG32 | Golestan/Gorgan |
| BRTRS | LR45 | Lorestan/Khoram Abad |
Wheat germplasm genotyping using DArTseq platform
A diversity panel of 185 wheat genotypes was grown in a controlled greenhouse. Young leaves from 10-day-old seedlings were used for DNA extraction following the protocol recommended by Diversity Array Technology (DArT) company and whole wheat genotypes were genotyped with the wheat DArTseq platform using the Pst1 complexity reduction method as described before71. DArTseq markers were filtered to retain markers with known chromosomal position, markers with ≤ 20% missing data and minor allele frequency (MAF) ≥ 5%72.
Genetic diversity, population structure and linkage disequilibrium (LD)
The genetic diversity and population structure of 185 wheat genotypes were previously described73. Briefly, Cluster analysis of diversity panel estimated in DARwin ver. 5.0 software using the Unweight Neighbor-Joining (UNJ) algorithm. Pairwise LD between markers was measured as r2 by plotting the r2 against the pairwise genetic distance between markers74,75. The graphical LD decay was imputed by the GAPIT R package76.
Population structure of the 185 wheat genotypes was performed in STRUCTURE 2.1 using the Bayesian clustering algorithm with a burn-in period at 10,000 interactions followed by 10,000 replication of Markov Chain Monte Carlo (MCMC)77.
Genome-wide association analysis for seedling leaf rust resistance
To meet the data format required for GWAS analysis, infection types (IT) data were converted into a linearized scale (LS) of 0–9 scale as described78. ITs were converted as follows: 0, 1−, 1, 1+, 2−, 2, 2+, 3−, 3 and 3+ were coded as 0, 1, 2, 3, 4, 5, 6, 7, 8 and 9, respectively. The IT symbol “;” and 4 converted to 0 and 9, respectively. Mesothetic reaction types X −, X, and X + were converted to linearized scores of 4, 5, and 6, respectively. The BLUE value of linearized scale (0–9) for all isolates was calculated using the PROC MIXED procedure in SAS v9.3. In the model, the genotype considered as a fixed effect and replication (block) considered as random effect. These BLUE values were then used for broad sense heritability estimates and correlations between isolates, cluster analysis of wheat genotypes and also to perform GWAS73. Genome-wide association mapping (GWAS) analysis was conducted in the R package Genome Association and Prediction Integrated Tool (GAPIT)76 using all 21,773 mapped polymorphic DArTseq markers.
Association analysis for each Pt race was conducted using the FarmCPU model35,79. Association results of the FarmCPU model were compared with association models like as GLM, MLM, CMLM and Super-MLM models and finally, this model provided a robust model for association mapping of resistance genes against Pt races, which effectively controls both false positives and false negatives80.
The quantile–quantile (Q-Q) plot of each Pt race was drawn using the observed and expected log10 P values. Marker–trait associations (MTAs) were selected if the significant markers cross the false discovery rate threshold (P = 0.05) and a uniform threshold level of P-value ≥ 0.0001 (− log10 P = 4.00). Significant MTAs associated with Pt races were ordered according to their genetic map positions in a high-resolution DArT-seq consensus map (version 4.0), provided by Dr. Andrzej Kilian (Diversity Arrays Technology Pty Ltd, Canberra, Australia). The identified QTLs and catalogued Lr genes81 were projected onto the wheat integrated consensus map38 and their positions were compared with previously known Lr genes and 393 QTLs from 50 QTL mapping studies39. Each QTL was considered new if its position was ≥ 10 cM from previously reported Lr genes or QTLs12. In order to find the candidate genes linked to MTAs, the physical position of these markers was taken to Ensembl using IWGSC RefSeq v1.0 genome and ~ 2.5 Mb flanking each marker was considered for annotated genes19.
Ethics approval and consent to participate
All the plant materials provided by Iranian Seed and Plant Improvement Institute (SPII) and were in compliance with relevant institutional, national, and international guidelines and legislation.
Supplementary Information
Acknowledgements
The authors acknowledge the Iranian Seed and Plant Improvement Institute (SPII) for their kind support in germplasm preparation and diseases phenotyping experiments.
Author contributions
R.T. and R.M. contributed to the material preparation, phenotyping, data collection & analysis, Writing—review & editing; M.M. contributed to the disease phenotyping, genotyping and data collection; A.M.N. contributed to the material preparation & data analysis.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
The plant materials used during the current study are available from the corresponding author on reasonable request. The DArTseq datasets generated and analyzed during the current study are available in the Figshare repository: https://figshare.com/articles/dataset/DArTseq-Data_xlsx/21967460.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Reza Talebi, Email: rezatalebi56@gmail.com.
Rahim Mehrabi, Email: mehrabi@iut.ac.ir.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-31559-y.
References
- 1.Zohary D, Hopf M, Weiss E. Domestication of Plants in the Old World: The origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean Basin. Oxford University Press on Demand; 2012. [Google Scholar]
- 2.Delfan S, Bihamta MR, Dadrezaei ST, Abbasi A, Alipoor H. Exploring genomic regions involved in bread wheat resistance to leaf rust at seedling/adult stages by using GWAS analysis. BMC Genom. 2023;24:83. doi: 10.1186/s12864-022-09096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nemati Z, Mostowfizadeh-Ghalamfarsa R, Dadkhodaie A, Mehrabi R, Steffenson BJ. Virulence of leaf rust physiological races in Iran from 2010 to 2017. Plant Dis. 2020;104(2):363–372. doi: 10.1094/PDIS-06-19-1340-RE. [DOI] [PubMed] [Google Scholar]
- 4.Kolmer JA. Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 2005;8:441–449. doi: 10.1016/j.pbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Rafiei-Boroujeni F, Arzani A, Afshari F, Torabi M. Postulation of leaf rust resistance genes in Iranian wheat cultivars and breeding lines. Can. J. Plant Pathol. 2011;33:550–558. doi: 10.1080/07060661.2011.630023. [DOI] [Google Scholar]
- 6.Safavi SA, Afshari F. Quantitative resistance of some Elite wheat lines to Puccinia striiformis f. sp. tritici. Arch. Phytopathol. Plant Prot. 2012;45:740–749. doi: 10.1080/03235408.2012.665216. [DOI] [Google Scholar]
- 7.Figlan S, Ntushelo K, Mwadzingeni L, Terefe T, Tsilo TJ, Shimelis H. Breeding wheat for durable leaf rust resistance in Southern Africa: Variability, distribution, current control strategies, challenges and future prospects. Front. Plant Sci. 2020;11:549. doi: 10.3389/fpls.2020.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoun M, Rouse MN, Kolmer JA, Kumar A, Elias EM. Genome-wide association studies reveal all-stage rust resistance loci in elite durum wheat genotypes. Front. Plant Sci. 2021;12:640739. doi: 10.3389/fpls.2021.640739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolmer JA, Liu JQ. Virulence and molecular polymorphism in international collections of the wheat leaf rust fungus Puccinia triticina. Phytopathology. 2000;90:427–436. doi: 10.1094/PHYTO.2000.90.4.427. [DOI] [PubMed] [Google Scholar]
- 10.Prasad P, Savadi S, Bhardwaj SC, Gangwar OP, Subodh KS. Rust pathogen effectors: Perspectives in resistance breeding. Planta. 2019;250:1–22. doi: 10.1007/s00425-019-03167-6. [DOI] [PubMed] [Google Scholar]
- 11.Ali S, Rodriguez-Algaba J, Thach T, Sørensen CK, Hansen JG, Lassen P, Nazari K, Hodson DP, Justesen AF, Hovmøller MS. Yellow rust epidemics worldwide were caused by pathogen races from divergent genetic lineages. Front. Plant Sci. 2017;8:1–14. doi: 10.3389/fpls.2017.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riaz A, Athiyannan N, Periyannan SK, Afanasenko O, Mitrofanova OP, Platz GJ, Aitken EAB, Snowdon RJ, Lagudah ES, Hickey LT, Voss-Fels KP. Unlocking new alleles for leaf rust resistance in the Vavilov wheat collection. Theor. Appl. Genet. 2018;131(1):127–144. doi: 10.1007/s00122-017-2990-5. [DOI] [PubMed] [Google Scholar]
- 13.Kolmer J, Hanzalova A, Goyeau H, Bayles R, Morgounov A. Genetic differentiation of the wheat leaf rust fungus Puccinia triticina in Europe. Plant Pathol. 2013;62:21–31. doi: 10.1111/j.1365-3059.2012.02626.x. [DOI] [Google Scholar]
- 14.Sapkota S, Hao Y, Johnson J, Buck J, Aoun M, Mergoum M. Genome-wide association study of a worldwide collection of wheat genotypes reveals novel quantitative trait loci for leaf rust resistance. Plant Genome. 2019;12:190033. doi: 10.3835/plantgenome2019.05.0033. [DOI] [PubMed] [Google Scholar]
- 15.Aoun M, Breiland M, Turner KM, Loladze A, Chao S, Xu SS, Ammar K, Anderson JA, Kolmer JA, Acevedo M. Genome-wide association mapping of leaf rust response in a durum wheat worldwide germplasm collection. Plant Genome. 2016;9:1–24. doi: 10.3835/plantgenome2016.01.0008. [DOI] [PubMed] [Google Scholar]
- 16.Flath K, Miedaner T, Olivera PD, Rouse MN, Yue J. Genes for wheat stem rust resistance postulated in German cultivars and their efficacy in seedling and adult-plant field tests. Plant Breed. 2018;137:301–312. doi: 10.1111/pbr.12591. [DOI] [Google Scholar]
- 17.Rahmatov M, Otambekova M, Muminjanov H, Rouse MN, Hovmøller MS, Nazari K, Steffenson BJ, Eva JE. Characterization of stem, stripe and leaf rust resistance in Tajik bread wheat accessions. Euphytica. 2019;215:55. doi: 10.1007/s10681-019-2377-6. [DOI] [Google Scholar]
- 18.Casassola A, Brammer SP, Chaves MS, Martinelli JA, Stefanato F, Boyd LA. Changes in gene expression profiles as they relate to the adult plantleaf rust resistance in the wheat cv. Toropi. Physiol. Mol. Plant. Pathol. 2015;89:49–54. doi: 10.1016/j.pmpp.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juliana P, Singh RP, Singh PK, Poland JA, Bergstrom GC, Huerta-Espino J, Bhavani S, Crossa J, Sorrells ME. Genome-wide association mapping for resistance to leaf rust, stripe rust and tan spot in wheat reveals potential candidate genes. Theor. Appl. Genet. 2018;131:1405–1422. doi: 10.1007/s00122-018-3086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juliana P, Singh RP, Huerta-Espino J, Bhavani S, Randhawa MS, Kumar U, Joshi AK, Bhati PK, Mir HEV, Nath CN, Singh GP. Genome-wide mapping and allelic fngerprinting provide insights into the genetics of resistance to wheat stripe rust in India, Kenya and Mexico. Sci. Rep. 2020;10:10908. doi: 10.1038/s41598-020-67874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rollar S, Serfling A, Geyer M, Hartl L, Mohler V, Ordon F. QTL mapping of adult plant and seedling resistance to leaf rust (Puccinia triticina Eriks.) in a multiparent advanced generation intercross (MAGIC) wheat population. Theor. Appl. Genet. 2021;134:37–51. doi: 10.1007/s00122-020-03657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omrani A, Aharizad S, Roohparvar R, Khodarahmi M, Toorchi M. Identification of stem and leaf rust resistance genes in some promising wheat lines using molecular markers. Crop Biotech. 2017;7(18):15–25. [Google Scholar]
- 23.Delfan S, Bihamta MR, Dadrezaei ST, Abbasi A, Alipour H. Identification sources of resistance for leaf rust (Puccinia triticina Eriks.) in Iranian wheat germplasm. Iran. J. Plant Prot. Sci. 2022;52(2):115–133. [Google Scholar]
- 24.Guo Y, Betzen B, Salcedo A, He F, Bowden RL, Fellers JP, Jordan KW, Akhunova A, Rouse MN, Szabo LJ, Akhunov E. Population genomics of Puccinia graminis f.sp. tritici highlights the role of admixture in the origin of virulent wheat rust races. Nat. Commun. 2022;13:6287. doi: 10.1038/s41467-022-34050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czajowski G, Kosman E, Słowacki P, Park RF, Czembor P. Pathogenic and genetic diversity of Puccinia triticina from triticale in Poland between 2012 and 2015. Plant Pathol. 2021;70:2148–2164. doi: 10.1111/ppa.13450. [DOI] [Google Scholar]
- 26.Kolmer JA, Kabdulova MG, Mustafina MA, Zhemchuzhina NS, Dubovoy V. Russian populations of Puccinia triticina in distant regions are not differentiated for virulence and molecular genotype. Plant Pathol. 2015;64(2):328–336. doi: 10.1111/ppa.12248. [DOI] [Google Scholar]
- 27.Rattu AR, Ahmad I, Singh RP, Fayyaz M, Mirza JI, Khanzada KA, Haque MI. Resistance to Puccinia triticina in some Pakistani wheats. Pak. J. Bot. 2010;42:2719–2735. [Google Scholar]
- 28.Kolmer JA. Virulence of Puccinia triticina, the wheat leaf rust fungus, in the United States in 2017. Plant Dis. 2019;103(8):2113–2120. doi: 10.1094/PDIS-09-18-1638-SR. [DOI] [PubMed] [Google Scholar]
- 29.Sibikeev SN, Krupnov VA, Voronina SA, Elesin VA. First report of leaf rust pathotypes virulent to highly effective Lr genes transferred from Agropyron species to bread wheat. Plant Breed. 1996;115:276–278. doi: 10.1111/j.1439-0523.1996.tb00917.x. [DOI] [Google Scholar]
- 30.Al-Maaroof EM, Singh RP, Huerta J, Rattu A. Resistance of some Iraqi bread wheat cultivars to Puccinia triticina. Phytopathol. Mediterr. 2005;44:247–255. [Google Scholar]
- 31.Yahyaoui, A., Hakim, S., Al-Naimi, M. & Nachit, M. M. Multiple disease resistance in durum wheat (Triticum turgidum L. var durum). CIHEAM: Série A. Séminaires Méditerranéens40, 387–392 (2000).
- 32.Stuckenbrock EH, Banke S, Javan-Nikkhah M, McDonald BA. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol. Biol. Evol. 2007;24:398–411. doi: 10.1093/molbev/msl169. [DOI] [PubMed] [Google Scholar]
- 33.Liu W, Kolmer J, Rynearson S, Chen X, Gao L, Anderson JA, Turner MK, Pumphrey M. Identifying loci conferring resistance to leaf and stripe rusts in a spring wheat population (Triticum aestivum) via genome-wide association mapping. Phytopathology. 2019;109(11):1932–1940. doi: 10.1094/PHYTO-04-19-0143-R. [DOI] [PubMed] [Google Scholar]
- 34.Kanwal M, Qureshi M, Gessese M, Forrest K, Babu P, Bariana H, Bansal U. An adult plant stripe rust resistance gene maps on chromosome 7A of Australian wheat cultivar Axe. Theor. Appl. Genet. 2021;134:2213–2220. doi: 10.1007/s00122-021-03818-x. [DOI] [PubMed] [Google Scholar]
- 35.Ward BP, Merrill K, Bulli P, Pumphrey M, Mason RE, Mergoum M, Johnson J, Sapkota S, Lopez B, Marshall D, Brown-Guedira G. Analysis of the primary sources of quantitative adult plant resistance to stripe rust in U.S. soft red winter wheat germplasm. Plant Genome. 2021;14:e20082. doi: 10.1002/tpg2.20082. [DOI] [PubMed] [Google Scholar]
- 36.Liu W, Maccaferri M, Bulli P, Rynearson S, Tuberosa R, Chen X, Pumphery M. Genome-wide association mapping for seedling and field resistance to Puccinia striiformis f. sp. tritici in elite durum wheat. Theor. Appl. Genet. 2017;130:649–667. doi: 10.1007/s00122-016-2841-9. [DOI] [PubMed] [Google Scholar]
- 37.Desiderio F, Guerra D, Rubiales D, Piarulli L, Pasquini M, Mastrangelo A, Siméone R, Blanco A, Cattivelli L, Valè G. Identification and mapping of quantitative trait loci for leaf rust resistance derived from a tetraploid wheat Triticum dicoccum accession. Mol. Breed. 2014;34:1659–1675. doi: 10.1007/s11032-014-0186-0. [DOI] [Google Scholar]
- 38.Kumar D, Kumar A, Chhokar V, Gangwar OP, Bhardwaj SC, Sivasamy M, Prasad SVS, Prakasha TL, Khan H, Singh R, Sharma P, Sheoran S, Iquebal MA, Jaiswal S, Angadi UB, Singh G, Rai A, Singh GP, Kumar D, Tiwari R. Genome-Wide association studies in diverse spring wheat panel for stripe, stem, and leaf rust resistance. Front. Plant Sci. 2020;11:748. doi: 10.3389/fpls.2020.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, Bossolini E, Chen X, Pumphrey M, Dubcovsky J. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.) G3. 2015;5:449–465. doi: 10.1534/g3.114.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aduragbemi A, Soriano JM. Unravelling consensus genomic regions conferring leaf rust resistance in wheat via meta-QTL analysis. Plant Genome. 2022;15:e20185. doi: 10.1002/tpg2.20185. [DOI] [PubMed] [Google Scholar]
- 41.Azzimonti G, Marcel TC, Robert O, Paillard S, Lannou C, Goyeau H. Diversity, specificity and impacts on field epidemics of QTLs involved in components of quantitative resistance in the wheat leaf rust pathosystem. Mol. Breed. 2014;34:549–567. doi: 10.1007/s11032-014-0057-8. [DOI] [Google Scholar]
- 42.Zhou Y, Ren Y, Lillemo M, Yao Z, Zhang P, Xia X, He Z, Li Z, Liu D. QTL mapping of adult-plant resistance to leaf rust in a RIL population derived from a cross of wheat cultivars Shanghai 3/Catbird and Naxos. Theor. Appl. Genet. 2014;127:1873–1883. doi: 10.1007/s00122-014-2346-3. [DOI] [PubMed] [Google Scholar]
- 43.Lan C, Zhang Y, Herrera-Foessel SA, Basnet BR, Huerta-Espino J, Lagudah ES, Singh RP. Identification and characterization of pleiotropic and co-located resistance loci to leaf rust and stripe rust in bread wheat cultivar Sujata. Theor. Appl. Genet. 2015;128:549–561. doi: 10.1007/s00122-015-2454-8. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P, Yin G, Zhou Y, Qi A, Gao F, Xia X, He Z, Li Z, Liu D. QTL mapping of adult-plant resistance to leaf rust in the wheat cross Zhou 8425B/ Chinese Spring using high-density SNP markers. Front. Plant Sci. 2017;8:793. doi: 10.3389/fpls.2017.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang P, Lan C, Asad M, Gebrewahid TW, Xia X, He Z, Li Z, Liu D. QTL mapping of adult-plant resistance to leaf rust in the Chinese landraces Pingyuan 50/Mingxian 169 using the wheat 55K SNP array. Mol. Breed. 2019;39:1–14. doi: 10.1007/s11032-019-1004-5. [DOI] [Google Scholar]
- 46.Kthiri D, Loladze A, N’Diaye A, Nilsen KT, Walkowiak S, Dreisigacker S, Ammar K, Pozniak CJ. Mapping of genetic loci conferring resistance to leaf rust from three globally resistant durum wheat sources. Front. Plant Sci. 2019;10:1247. doi: 10.3389/fpls.2019.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosa SB, Zanella CM, Hiebert C, Babel AB, Randhawa H, Shorter S, Boyd L, Mccallum B. Genetic characterization of leaf and stripe rust resistance in the Brazilian wheat cultivar Toropi. Phytopathology. 2019;109(10):1760–1768. doi: 10.1094/PHYTO-05-19-0159-R. [DOI] [PubMed] [Google Scholar]
- 48.Shewabez E, Bekele E, Alemu A, Mugnai L, Tadesse W. Genetic characterization and genome-wide association mapping for stem rust resistance in spring bread wheat. BMC Genom. Data. 2022;23:11. doi: 10.1186/s12863-022-01030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Dundas I, Dong C, Li G, Trethowan R, Yang Z, Hoxha S, Zhang P. Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 2020;133(4):1095–1107. doi: 10.1007/s00122-020-03534-y. [DOI] [PubMed] [Google Scholar]
- 50.Kertho A, Mamidi S, Bonman JM, McClean PE, Acevedo M. Genome-wide association mapping for resistance to leaf and stripe rust in winter-habit hexaploid wheat landraces. PLoS ONE. 2015;10:e0129580. doi: 10.1371/journal.pone.0129580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maccaferri M, Mantovani P, Tuberosa R, Deambrogio E, Giuliani S, Demontis A, Massi A, Sanguineti M. A major QTL for durable leaf rust resistance widely exploited in durum wheat breeding programs maps on the distal region of chromosome arm 7BL. Theor. Appl. Genet. 2008;117:1225–1240. doi: 10.1007/s00122-008-0857-5. [DOI] [PubMed] [Google Scholar]
- 52.Bokore FE, Knox R, Cuthbert R, Pozniak C, Mccallum B, N'Diaye A, DePauw R, Campbell H, Munro C, Singh A, Hiebert C, McCartney C, Sharpe A, Singh A, Spaner D, Fowler DB, Ruan Y, Berraies S, Meyer B. Mapping quantitative trait loci associated with leaf rust resistance in five spring wheat populations using single nucleotide polymorphism markers. PLoS ONE. 2020;15(4):e0230855. doi: 10.1371/journal.pone.0230855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh A, Knox R, DePauw R, Cuthbert R, Campbell H, Shorter S, Bhavani S. Stripe rust and leaf rust resistance QTL mapping epistatic interactions and co-localization with stem rust resistance loci in spring wheat evaluated over three continents. Theor. Appl. Genet. 2014;127:2465–2477. doi: 10.1007/s00122-014-2390-z. [DOI] [PubMed] [Google Scholar]
- 54.Ren Y, Li Z, He Z, Wu L, Bai B, Lan C, Wang C, Zhou G, Zhu H, Xia X. QTL mapping of adult-plant resistances to stripe rust and leaf rust in Chinese wheat cultivar Bainong 64. Theor. Appl. Genet. 2012;125:1253–1262. doi: 10.1007/s00122-012-1910-y. [DOI] [PubMed] [Google Scholar]
- 55.Chu CG, Friesen TL, Xu SS, Faris JD, Kolmer JA. Identification of novel QTLs for seedling and adult plant leaf rust resistance in a wheat doubled haploid population. Theor. Appl. Genet. 2009;119:263–269. doi: 10.1007/s00122-009-1035-0. [DOI] [PubMed] [Google Scholar]
- 56.Lan CX, Rosewarne GM, Singh RP, Herrera-Foessel SA, HuertaEspino J, Basnet BR, Zhang YL, Yang EN. QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin#1. Mol. Breed. 2014;34:789–803. doi: 10.1007/s11032-014-0075-6. [DOI] [Google Scholar]
- 57.Faris JD, Li WL, Liu DJ, Chen PD, Gill BS. Candidate gene analysis of quantitative disease resistance in wheat. Theor. Appl. Genet. 1999;98:219–225. doi: 10.1007/s001220051061. [DOI] [Google Scholar]
- 58.Genievskaya Y, Abugalieva S, Rsaliyev A, Yskakova G, Turuspekov Y. QTL mapping for seedling and adult plant resistance to leaf and stem rusts in Pamyati Azieva × Paragon mapping population of bread wheat. Agronomy. 2020;10(9):1285. doi: 10.3390/agronomy10091285. [DOI] [Google Scholar]
- 59.Lan C, Basnet BR, Singh R, Huerta-Espino J, Herrera-Foessel S, Ren Y, Randhawa M. Genetic analysis and mapping of adult plant resistance loci to leaf rust in durum wheat cultivar Bairds. Theor. Appl. Genet. 2017;130:609–619. doi: 10.1007/s00122-016-2839-3. [DOI] [PubMed] [Google Scholar]
- 60.Li C, Wang Z, Li C, Bowden R, Bai G, Li C, Su Z, Carver B. Mapping of quantitative trait loci for leaf rust resistance in the wheat population Ning7840 × Clark. Plant Dis. 2017;101(12):1974–1979. doi: 10.1094/PDIS-12-16-1743-RE. [DOI] [PubMed] [Google Scholar]
- 61.Dubcovsky J, Lukaszewski AJ, Echaide M, Antonelli EF, Porter DR. Molecular characterization of two Triticum speltoides interstitial translocations carrying leaf rust and greenbug resistance genes. Crop Sci. 1998;38:1655–1660. doi: 10.2135/cropsci1998.0011183X003800060040x. [DOI] [Google Scholar]
- 62.Lu Y, Bowden R, Zhang G, Xu X, Fritz A, Bai G. Quantitative trait loci for slow-rusting resistance to leaf rust in doubled-haploid wheat population CI13227 × Lakin. Phytopathology. 2017;107(11):1372–1380. doi: 10.1094/PHYTO-09-16-0347-R. [DOI] [PubMed] [Google Scholar]
- 63.Qi A, Zhang P, Zhou Y, Yao Z, Zai-feng L, Liu D. Mapping of QTL conferring leaf rust resistance in Chinese wheat lines W014204 and Fuyu 3 at adult plant stage. J. Integr. Agric. 2016;15:18–28. doi: 10.1016/S2095-3119(14)60974-6. [DOI] [Google Scholar]
- 64.Bemister DH, Semagn K, Iqbal M, Randhawa H, Strelkov S, Spaner D. Mapping QTL associated with stripe rust leaf rust and leaf spotting in a Canadian spring wheat population. Crop Sci. 2019;59:650–658. doi: 10.2135/cropsci2018.05.0348. [DOI] [Google Scholar]
- 65.Long DL, Kolmer JA. A North American system of nomenclature for Puccinia recodita f. sp. tritici. Phytopathology. 1989;79:525–529. doi: 10.1094/Phyto-79-525. [DOI] [Google Scholar]
- 66.Kolmer JA, Long DL, Hughes ME. Physiological specialization of Puccinia triticina on wheat in the United States in 2005. Plant Dis. 2007;91:979–984. doi: 10.1094/PDIS-91-8-0979. [DOI] [PubMed] [Google Scholar]
- 67.Singh RP, Rajaram S. Resistance to Puccinia recondite f.sp. tritici in 50 Mexican bread wheat cultivars. Crop Sci. 1991;31:1472–1479. doi: 10.2135/cropsci1991.0011183X003100060016x. [DOI] [Google Scholar]
- 68.Stakman EC, Stewart DM, Loegering WQ. Identification of Physiologic Races of Puccinia graminis var. Tritici. 1962; USDA, Washington, DC.
- 69.Huerta-Espino J, Singh R, Herrera-Foessel S, Perez-Lopez J, Figueroa-Lopez P. First detection of virulence in Puccinia triticina to resistance genes Lr27+Lr31 present in durum wheat in Mexico. Plant Dis. 2009;93:110. doi: 10.1094/PDIS-93-1-0110C. [DOI] [PubMed] [Google Scholar]
- 70.Hammer O, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Paleontol. Electron. 2001;4:1–9. [Google Scholar]
- 71.Egea LA, Mérida-García R, Kilian A, Hernandez P, Dorado G. Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by Diversity Arrays Technology “genotyping-by-sequencing” platform (DArTseq) Front. Genet. 2017;8:98. doi: 10.3389/fgene.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping ofcomplex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 73.Mahboubi M, Talebi R, Mehrabi R, Naji AM, Maccaferri M, Kema G. Genetic analysis of novel resistance sources and genome-wide association mapping identified novel QTLs for resistance to Zymoseptoria tritici, the causal agent of septoria tritici blotch in wheat. J. Appl. Genet. 2022;63(3):429–445. doi: 10.1007/s13353-022-00696-x. [DOI] [PubMed] [Google Scholar]
- 74.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, International Wheat Genome Sequencing Consortium. Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E. Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014;12(6):787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nielsen NH, Backes G, Stougaard J, Andersen SU, Jahoor A. Genetic diversity and population structure analysis of European hexaploid bread wheat (Triticum aestivum L.) varieties. PLoS ONE. 2014;9:e94000. doi: 10.1371/journal.pone.0094000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics. 2012;28(18):2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 77.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters ofindividuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 78.Zhang D, Bowden RL, Yu J, Carver BF, Bai G. Association analysis of stem rust resistance in U.S. winter wheat. PLoS ONE. 2014;9(7):e103747. doi: 10.1371/journal.pone.0103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaler AS, Gillman JD, Beissinger T, Purcell LC. Comparing different statistical models and multiple testing corrections for association mapping in soybean and maize. Front. Plant Sci. 2020;10:1794. doi: 10.3389/fpls.2019.01794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y, Qie Y, Li X, Wang M, Chen X. Genome-wide mapping of quantitative trait loci conferring all-stage and high-temperature adult-plant resistance to stripe rust in spring wheat landrace PI1840. Int. J. Mol. Sci. 2020;21:1594. doi: 10.3390/ijms21020478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Xia XC. Catalogue of Gene Symbols for Wheat: 2017; Supplement. https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2017.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plant materials used during the current study are available from the corresponding author on reasonable request. The DArTseq datasets generated and analyzed during the current study are available in the Figshare repository: https://figshare.com/articles/dataset/DArTseq-Data_xlsx/21967460.