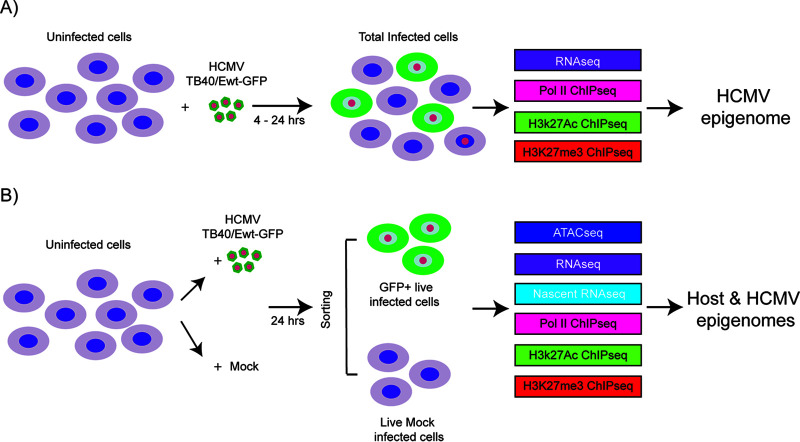
FIG 1.
Schematic of the samples used in the analysis of host and HCMV transcriptomes and epigenomes in Kasumi-3 cells. (A) Kasumi-3 cells were infected with HCMV TB40/Ewt-GFP at an MOI of 1. The total cell population, which includes GFP+ and GFP− infected cells as well as uninfected cells, was collected at 4 and 24 hpi. Cells were used for RNA-seq, and Pol II, H3K27Ac, and H3K27me3 ChIP-seq analyses to study the HCMV epigenome. (B) Kasumi-3 cells were mock infected or infected with HCMV TB40/Ewt-GFP at an MOI of 1. At 24 hpi, cells were sorted to obtain GFP+ live infected cells and live mock-infected cells. Those populations were then subjected to ATAC-seq, RNA-seq, nascent RNA-seq, and Pol II, H3K27Ac and H3K27me3 ChIP-seq analyses to study the effect of HCMV infection on both host and viral epigenomes.