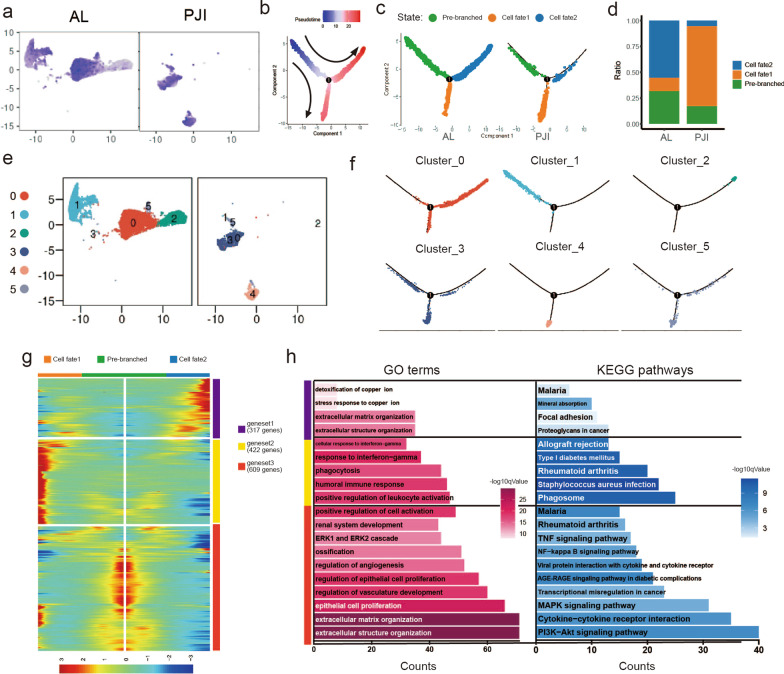
Fig. 5.
In-depth fibroblast trajectory analysis reveals a bipolar mode of differentiation. a UMAP visualization of the fibroblast, cells are colored according to CTHRC1 expression level. b Pseudotime trajectory analysis shows fibroblasts differentiation, cells are colored by pseudotime. c Trajectory plots show fibroblasts differentiation in AL and PJI groups, cells are colored by cell states. d Stacked bar plot showing proportions of each cell state in AL and PJI groups. e UMAP visualization of the fibroblast, cells are colored according to subclusters. f Fibroblast subclusters are projected to pseudotime trajectory, cells are colored by subclusters. g Heatmap revealed pseudotime-dependent differentially expressed gene clusters for cell fate1, cell fate2, and pre-branched cells. h Bar plots show the enrichment result of gene ontology and KEGG pathway for each of the gene clusters identified in (g), both are colored with log10qvalues