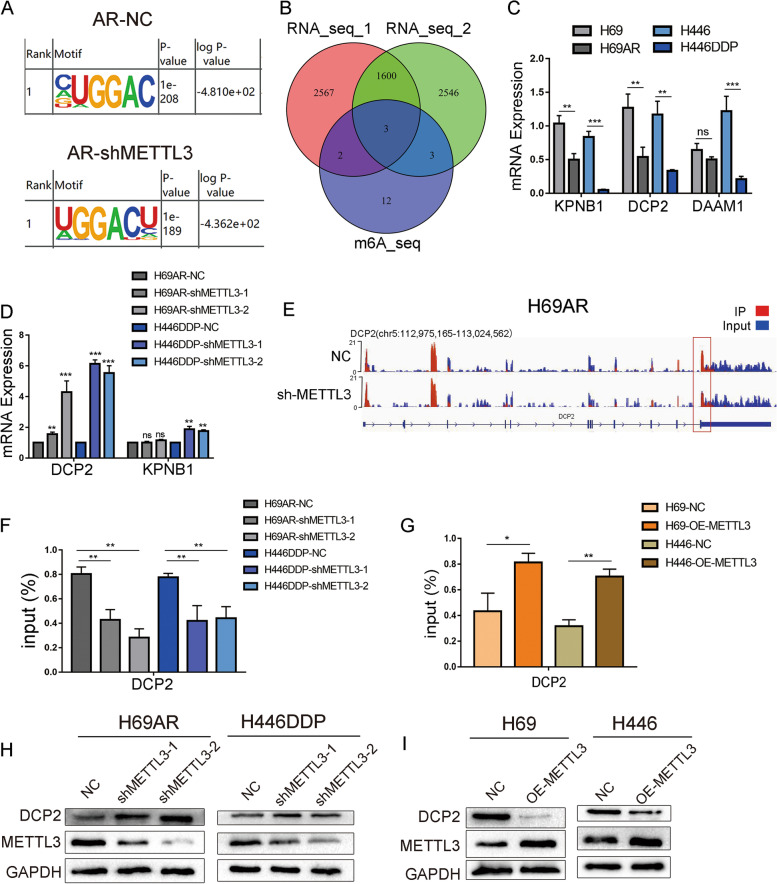
Fig. 4.
DCP2 is a downstream target of METTL3. A Consensus motif map with MeRIP-seq peaks identified by HOMER in the SCLC drug-resistant cell line H69AR after METTL3 knockdown. B Venn diagram showing the gene set with significantly decreased m6A levels following METTL3 knockdown as measured by MeRIP-seq and the two gene sets with significant differences in RNA-seq differential expression analysis. The intersection of these gene sets yielded three significantly different genes (DCP2, DAAM1, and KPNB1). C qRT-PCR analysis: mRNA expression of DCP2, DAAM1 and KPNB1 in sensitive and resistant cell lines was detected by qRT–PCR. **P < 0.01; ***P < 0.001; ns, not significant. D qRT-PCR analysis: mRNA expression of DCP2 and KPNB1 after METTL3 knockdown in the drug-resistant cell lines H69AR and H446DDP. **P < 0.01; ***P < 0.001; ns, not significant. E IGV map from MeRIP-seq shows that DCP2 m6A methylation levels were significantly decreased in METTL3-knockdown H69AR cells. F MeRIP-qPCR analysis of DCP2 m6A methylation levels in drug-resistant cell lines and their METTL3-knockdown stably transduced counterparts. **P < 0.01. G MeRIP-qPCR analysis of DCP2 m6A methylation levels in chemosensitive cell lines and their counterparts that stably overexpress METTL3. *P < 0.05, **P < 0.01. H Western blot analysis of DCP2 expression in H69AR and H446DDP cells transfected with shRNA-METTL3. I Western blot analysis of DCP2 expression in H69 and H446 cells transfected with METTL3