Abstract
Background:
Rifampicin (RIF) resistance in Mycobacterium tuberculosis is frequently caused by mutations in the rpoB gene. These mutations are associated with a fitness cost, which can be overcome by compensatory mutations in other genes, among which rpoC may be the most important. We analyzed 469 Peruvian M. tuberculosis clinical isolates to identify compensatory mutations in rpoC/rpoA associated with RIF resistance.
Methods:
The M. tuberculosis isolates were collected and tested for RIF susceptibility and spoligotyping. Samples were sequenced and aligned to the reference genome to identify mutations. By analyzing the sequences and the metadata, we identified a list of rpoC mutations exclusively associated with RIF resistance and mutations in rpoB. We then evaluated the distribution of these mutations along file protein sequence and tridimensional structure.
Results:
One hundred and twenty-five strains were RIF susceptible and 346 were resistant. We identified 35 potential new compensatory mutations, some of which were distributed on the interface surface between rpoB and rpoC, arising in clusters and suggesting the presence of hotspots for compensatory mutations.
Conclusion:
This study identifies 35 putative novel compensatory mutations in the β’ subunit of M. tuberculosis RNApol. Six of these (S428T, L507V, A734V, I997V, and V1252LM) are considered most likely to have a compensatory role, as they fall in the interaction zone of the two subunits and the mutation did not lead to any change in the protein’s physical–chemical properties.
Keywords: Compensatory mutations, evolution, rifampicin resistance, rpoB, rpoC, tuberculosi
Introduction
Tuberculosis (TB) is the leading cause of death by a single infectious agent worldwide.[1] With an average of 27,000 new cases per year, Peru has the second highest rate of TB in the Americas; thanks to a national TB control program,[2] the incidence of TB has decreased since 2000 by 1.6% per year.[3]
Rifampicin (RIF) is one of the most important drugs in the treatment of TB, and resistance to RIF threatens the success of TB control programs globally.[4] In 2017, the WHO reported more than 159,000 cases of RIF-resistant TB including 1,508 in Peru.[5] In the same year, about 390,000 new cases of multidrug-resistant TB (MDR-TB) were reported worldwide.[3] In the preceding decade, the average number of reported cases per year in Peru exceeded 1,100, with a trend toward an increase in the later years.[6]
RIF causes bacterial cell death by binding to the β-subunit of the bacteria’s RNA polymerase (RNApol), blocking RNA transcription, and inhibiting gene expression.[7-10] Mutations in rpoB, the gene encoding the Mycobacterium tuberculosis RNApol β-subunit, have been shown to be the main cause of RIF-resistant microorganisms.[11] Moreover, the RIF resistance-determining region (RRDR) is an 81 bp region in rpoB within which 95% of all known RIF-resistant mutations are found.[11] Many studies have examined the structural variations of rpoB mutants and their association with drug binding.[12] Escherichia coli, distinct species of Salmonella, Thermus, and Mycobacterium have been the most studied models for RIF resistance to date.
Mutations in rpoB have been found to be associated with reduced fitness of the affected strain. Several studies have demonstrated that the fitness loss due to rpoB mutations can be compensated for by other mutations, which may consequently be positively selected. These mutations have been mainly identified in the rpoC and/or rpoA genes which are responsible for coding the β’ and α-subunits of the RNApol complex, respectively, and which interact closely with rpoB.[8,10,13-15] Several compensatory[13,14,16-19] and putative compensatory[20-23] mutations in these two genes have been described, mostly in rpoC, and including the V483A and L516P mutations that have been extensively described in M. tuberculosis.[16,19,20,23]
In this study, we analyzed rpoA, rpoB, and rpoC gene sequences from 469M. tuberculosis clinical strains, recovered from unrelated TB patients from Peru, to identify compensatory mutations in rpoC/rpoA genes associated with RIF resistance. Characterization of these mutations will improve our understanding of the mycobacterial compensatory mechanisms that allow them to overcome the fitness cost associated with rpoB mutations and thereby enable the survival and ongoing transmission of RIF-resistant strains. Furthermore, this study could provide insight into evolutionary processes, leading to RIF resistance in TB.
Methods
Selection of strains and rifampicin-susceptibility testing
We selected 469 well-characterized MTB strains, isolated from unrelated TB patients in hospitals from Callao and South Lima, Peru, between 2009 and 2013. These were collected and selected as part of two large studies that have been described previously:[24] one was a population-level study and the other a household follow-up study. The MODS assay was used to confirm TB infection and determine RIF susceptibility. MODS is a highly sensitive (98%) and specific (98.6%) tool that can both diagnose TB and detect drug resistance by determining the presence or absence of bacterial growth under a fixed drug concentration, and is approved for routine clinical use.[24-26]
The genotypes of these isolates have previously been studied using spoligotyping,[24] which characterizes mycobacterial strains according to their hybridization patterns.[24,27] Thus, in this study, we also evaluated the association between RIF resistance and the spoligotype.
Sequences and mutations
The rpoA, rpoB, and rpoC gene sequences, and mutations in these genes compared to the wild-type reference strain H37Rv (GeneBank: NC_000962) were obtained from the complete genome sequences attained previously.[24] Briefly, samples were sequenced on the Illumina MiSeq platform (150 bp paired-end reads) and aligned to the reference H37Rv genome using standard methodology.
Mutations in rpoA and rpoC associated with rifampicin-resistant strains
The 469 strains were classified as RIF susceptible or RIF resistant according to their MODS result. Each group was subclassified based on the presence or absence of at least one mutation in the rpoB gene. Each of these four groups was further subclassified based on the presence or absence one or more mutations in rpoA or rpoC.
To identify possible compensatory mutations in rpoC, we selected the RIF-susceptible strains with mutations in rpoC and a wild-type rpoB gene, regardless of the presence or absence of mutations in rpoA (set A). We also selected the RIF-resistant strains harboring mutations in rpoC and any mutation in rpoB, regardless of rpoA status (set B). After removing all rpoC mutations from set B that were also present in set A, we obtained a list of rpoC mutations exclusively associated with RIF resistance and mutations in rpoB. The same approach was applied to mutations in rpoA.
rpoA and rpoC compensatory mutations reported elsewhere
A literature search was performed to identify any mutations in rpoC or rpoA that have previously been associated with RIF resistance and rpoB mutations and have been shown experimentally to be compensating for a loss of fitness.[13,14,16-23] These were compiled into a list, based on which we were able to classify the mutations identified in this study as either previously reported or as potentially novel.
Distribution of potentially novel compensatory rpoC mutations along the protein sequence
The distribution of rpoC mutations was represented in a linear histogram, which indicated an equitable amino acid spread along the complete sequence of the protein. The β’ subunit (coded by rpoC) sequence comprises 1,300 amino acids, and the histogram was built on blocks of 100 amino acids. Previously reported rpoC mutations and mutations identified in this study were mapped separately. Peruvian mutations that had already been reported elsewhere were marked. This analysis was not performed for mutations in rpoA because of insufficient mutations, either previously reported or identified in this study.
Distribution of potentially compensatory rpoC mutations on the three-dimensional structure of Mycobacterium tuberculosis RNA polymerase
To better understand the mechanism of action of the potentially compensatory rpoC mutations, these were mapped onto the recently published crystal structure of the M. tuberculosis RNApol transcription initiation complex[28] (PDB ID: 5UHD). The structures were visualized and analyzed using VMD.[29] First, all of the putative compensatory rpoC mutations were mapped onto the structure. Then, the mutations unique to this study were differentiated from those reported previously.
We visualized the distribution of the mutations in the β’ subunit (coded by rpoC) that fell within the region of the surface that interacts with the β subunit (coded by rpoB). To identify the residues of physical importance in the β subunit, we identified those that arise within 5.0–6.0 Å of these rpoC mutations.
Results
Determination of mutations in rpoB, rpoA, and rpoC
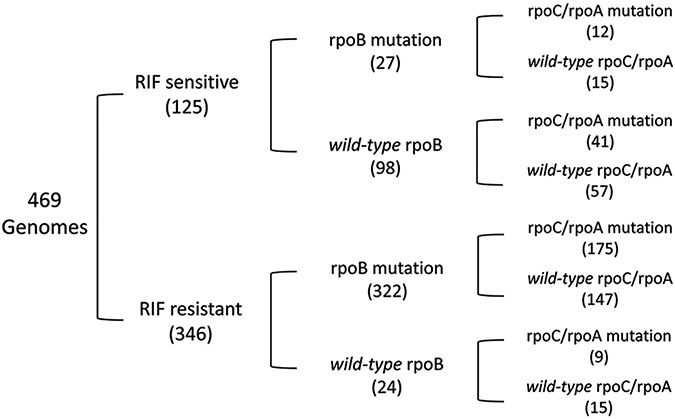
Among the 469 genomes analyzed [Table 1], we found 125 (27%) to be RIF sensitive and 346 (74%) to be RIF resistant according to the MODS assay [Figure 1]. We schematically distributed the strains according to their sensitivity to RIF, mutations in rpoB, and mutations in rpoC/rpoA [Figure 1]. 237 (50.5%) strains harbored mutation (s) in rpoC and/or rpoA. 322 (93.1%) RIF-resistant strains harbored at least one mutation in rpoB.
Table 1:
General database
| Strain number | rpoA | rpoC | rpoB | Spoligotyping | RRDR | RIF resistance |
|---|---|---|---|---|---|---|
| 14722_6#1_H37Rv | V483A | S450L | T | 1 | 1 | |
| 14722_6#10_H37Rv | LAM | 0 | 1 | |||
| 14722_6#11_H37Rv | H445D | Beijing | 1 | 1 | ||
| 14722_6#12_H37Rv | P1040R | S450L | Beijing | 1 | 1 | |
| 14722_6#13_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_6#14_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#15_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_6#16_H37Rv | I491V | S450L, A692T | LAM | 1 | 1 | |
| 14722_6#17_H37Rv | G594E | L452P | Haarlem | 1 | 0 | |
| 14722_6#18_H37Rv | LAM | 0 | 0 | |||
| 14722_6#19_H37Rv | LAM | 0 | 0 | |||
| 14722_6#2_H37Rv | I491V | S450L, A692T | LAM | 1 | 1 | |
| 14722_6#20_H37Rv | Bovis | 0 | 0 | |||
| 14722_6#21_H37Rv | G594E | E250G | Haarlem | 0 | 0 | |
| 14722_6#22_H37Rv | I965V | Caprae | 0 | 0 | ||
| 14722_6#23_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_6#24_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#25_H37Rv | LAM | 0 | 1 | |||
| 14722_6#26_H37Rv | LAM | 0 | 0 | |||
| 14722_6#27_H37Rv | Beijing | 0 | 0 | |||
| 14722_6#28_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_6#29_H37Rv | W484S | S450L | Beijing | 1 | 1 | |
| 14722_6#3_H37Rv | G594E | S450L | Other small clades | 1 | 1 | |
| 14722_6#30_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_6#31_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_6#32_H37Rv | T | 0 | 0 | |||
| 14722_6#33_H37Rv | R741S | H445C | LAM | 1 | 1 | |
| 14722_6#34_H37Rv | G594E | S450L, M920V | Haarlem | 1 | 1 | |
| 14722_6#35_H37Rv | LAM | 0 | 0 | |||
| 14722_6#36_H37Rv | D435F | Beijing | 1 | 0 | ||
| 14722_6#37_H37Rv | G594E | E250G | Haarlem | 0 | 0 | |
| 14722_6#38_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_6#39_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_6#4_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 14722_6#40_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#41_H37Rv | D435F | Beijing | 1 | 0 | ||
| 14722_6#42_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_6#43_H37Rv | G594E | H445Y | Other small clades | 1 | 1 | |
| 14722_6#44_H37Rv | Haarlem | 0 | 1 | |||
| 14722_6#45_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14722_6#46_H37Rv | S450L | Beijing | 1 | 1 | ||
| 14722_6#47_H37Rv | LAM | 0 | 1 | |||
| 14722_6#48_H37Rv | I965V | Caprae | 0 | 0 | ||
| 14722_6#49_H37Rv | T | 0 | 0 | |||
| 14722_6#5_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_6#50_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#51_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14722_6#52_H37Rv | S428G, Q432E, S672F | LAM | 1 | 1 | ||
| 14722_6#53_H37Rv | D435V | LAM | 1 | 0 | ||
| 14722_6#54_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#55_H37Rv | I491V G594E | S450L | Other small clades | 1 | 1 | |
| 14722_6#56_H37Rv | G594E | D435V | Other small clades | 1 | 1 | |
| 14722_6#57_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_6#58_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_6#59_H37Rv | L527V | S450L, V695L | T | 1 | 1 | |
| 14722_6#6_H37Rv | LAM | 0 | 0 | |||
| 14722_6#60_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#61_H37Rv | G594E, E1113D | I491F | Other small clades | 0 | 0 | |
| 14722_6#62_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 14722_6#63_H37Rv | G594E | S450L | Haarlem | 1 | 1 | |
| 14722_6#64_H37Rv | G594E, E1113D | I491F | Other small clades | 0 | 0 | |
| 14722_6#65_H37Rv | H445P, V695L | T | 1 | 1 | ||
| 14722_6#66_H37Rv | LAM | 0 | 0 | |||
| 14722_6#67_H37Rv | T812I | S450L, A692T | LAM | 1 | 1 | |
| 14722_6#68_H37Rv | LAM | 0 | 0 | |||
| 14722_6#69_H37Rv | LAM | 0 | 0 | |||
| 14722_6#7_H37Rv | V695L | T | 1 | 1 | ||
| 14722_6#70_H37Rv | Beijing | 0 | 0 | |||
| 14722_6#71_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_6#72_H37Rv | G594E | D435V | Haarlem | 1 | 1 | |
| 14722_6#73_H37Rv | G594E | S450F | Haarlem | 1 | 1 | |
| 14722_6#74_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_6#75_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#76_H37Rv | LAM | 0 | 0 | |||
| 14722_6#77_H37Rv | P45L, S450L | LAM | 1 | 1 | ||
| 14722_6#78_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_6#79_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_6#8_H37Rv | D435V | LAM | 0 | 0 | ||
| 14722_6#80_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#81_H37Rv | Beijing | 0 | 0 | |||
| 14722_6#82_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_6#83_H37Rv | T | 0 | 0 | |||
| 14722_6#84_H37Rv | V695L | T | 0 | 0 | ||
| 14722_6#85_H37Rv | Q980R | LAM | 1 | 1 | ||
| 14722_6#86_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_6#87_H37Rv | I491V | S450L, A692T | LAM | 1 | 1 | |
| 14722_6#88_H37Rv | I491V | S450L, A692T | LAM | 1 | 1 | |
| 14722_6#89_H37Rv | G594E | D435V | Haarlem | 1 | 1 | |
| 14722_6#9_H37Rv | L527V, G594E | S450L | Other small clades | 1 | 1 | |
| 14722_6#90_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_6#91_H37Rv | G594E | S450L | Haarlem | 1 | 1 | |
| 14722_6#92_H37Rv | T | 0 | 0 | |||
| 14722_6#93_H37Rv | LAM | 0 | 0 | |||
| 14722_6#94_H37Rv | G594E | S450F | Haarlem | 1 | 1 | |
| 14722_6#95_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#1_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_7#10_H37Rv | H525Q | S450L | Beijing | 1 | 1 | |
| 14722_7#11_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#12_H37Rv | R741S | S450L, D574E | LAM | 1 | 1 | |
| 14722_7#13_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_7#14_H37Rv | E207K, H445Y | LAM | 1 | 1 | ||
| 14722_7#15_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#16_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#17_H37Rv | W484S | S450L | Beijing | 1 | 1 | |
| 14722_7#18_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#19_H37Rv | V483A | S450L | LAM | 1 | 1 | |
| 14722_7#2_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#20_H37Rv | L527V | S450L | T | 1 | 1 | |
| 14722_7#21_H37Rv | G332R | S450L, V695L | T | 1 | 1 | |
| 14722_7#22_H37Rv | G594E | D435V | Other small clades | 1 | 1 | |
| 14722_7#23_H37Rv | E488G, G594E | S450L | Haarlem | 1 | 1 | |
| 14722_7#24_H37Rv | P1040R | S450L | LAM | 1 | 1 | |
| 14722_7#25_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#26_H37Rv | G594E, E744G | S450L | Other small clades | 1 | 1 | |
| 14722_7#27_H37Rv | Beijing | 0 | 1 | |||
| 14722_7#28_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#29_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#3_H37Rv | D435Y, N437H, V695L | T | 1 | 1 | ||
| 14722_7#30_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14722_7#31_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#32_H37Rv | H445L | LAM | 1 | 1 | ||
| 14722_7#33_H37Rv | LAM | 0 | 0 | |||
| 14722_7#34_H37Rv | P1040Q | S450L, V695L | T | 1 | 1 | |
| 14722_7#35_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#36_H37Rv | H445Y | LAM | 1 | 1 | ||
| 14722_7#37_H37Rv | G594E | P45S, S450L | Haarlem | 1 | 1 | |
| 14722_7#38_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 14722_7#39_H37Rv | T187A | S450L | Beijing | 1 | 1 | |
| 14722_7#4_H37Rv | LAM | 0 | 1 | |||
| 14722_7#40_H37Rv | S450W | T | 1 | 1 | ||
| 14722_7#41_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_7#42_H37Rv | G594E | A286V, S450L | Haarlem | 1 | 1 | |
| 14722_7#43_H37Rv | LAM | 0 | 0 | |||
| 14722_7#44_H37Rv | V1252L | S450L, V695L | T | 1 | 1 | |
| 14722_7#45_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#46_H37Rv | G594E | Haarlem | 0 | 1 | ||
| 14722_7#47_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#48_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#49_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_7#5_H37Rv | LAM | 0 | 1 | |||
| 14722_7#50_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14722_7#51_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#52_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_7#53_H37Rv | G594E, V1147A | S450L | Other small clades | 1 | 1 | |
| 14722_7#54_H37Rv | LAM | 0 | 0 | |||
| 14722_7#55_H37Rv | LAM | 0 | 0 | |||
| 14722_7#56_H37Rv | LAM | 0 | 0 | |||
| 14722_7#57_H37Rv | LAM | 0 | 0 | |||
| 14722_7#58_H37Rv | G31S | G594E | S450L | Other small clades | 1 | 1 |
| 14722_7#59_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#6_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#60_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#61_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_7#62_H37Rv | Q523E | S450L | Beijing | 1 | 1 | |
| 14722_7#63_H37Rv | L516Q | S450L | LAM | 1 | 1 | |
| 14722_7#64_H37Rv | LAM | 0 | 0 | |||
| 14722_7#65_H37Rv | T812I | S450L, A692T | LAM | 1 | 1 | |
| 14722_7#66_H37Rv | G31S | G594E | S450L | Other small clades | 1 | 1 |
| 14722_7#67_H37Rv | R741S | LAM | 0 | 1 | ||
| 14722_7#68_H37Rv | LAM | 0 | 0 | |||
| 14722_7#69_H37Rv | LAM | 0 | 0 | |||
| 14722_7#7_H37Rv | G571R, G594E | V170F | Other small clades | 0 | 1 | |
| 14722_7#70_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_7#71_H37Rv | LAM | 0 | 0 | |||
| 14722_7#72_H37Rv | H445D, V695L | T | 1 | 1 | ||
| 14722_7#73_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#74_H37Rv | G594E | E250G, S450L, F971L | Haarlem | 1 | 1 | |
| 14722_7#75_H37Rv | G594E | L452P | Haarlem | 1 | 0 | |
| 14722_7#76_H37Rv | G594E, F831L | S450L | Other small clades | 1 | 1 | |
| 14722_7#77_H37Rv | S450L, A599V, V695L | T | 0 | 1 | ||
| 14722_7#78_H37Rv | L1245R | S450L | Beijing | 1 | 1 | |
| 14722_7#79_H37Rv | LAM | 0 | 0 | |||
| 14722_7#8_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 14722_7#80_H37Rv | LAM | 0 | 0 | |||
| 14722_7#81_H37Rv | G594E | H445Y | Haarlem | 1 | 1 | |
| 14722_7#82_H37Rv | G332R | S450L, V695L | T | 1 | 1 | |
| 14722_7#83_H37Rv | G594E | S450L, M920V | Haarlem | 1 | 1 | |
| 14722_7#84_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_7#85_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14722_7#86_H37Rv | S450L, Q980R | LAM | 1 | 1 | ||
| 14722_7#87_H37Rv | G332R | S450L, V695L | T | 1 | 1 | |
| 14722_7#88_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#89_H37Rv | LAM | 0 | 0 | |||
| 14722_7#9_H37Rv | S450L | Beijing | 1 | 1 | ||
| 14722_7#90_H37Rv | G594E | S450L | Other small clades | 1 | 1 | |
| 14722_7#91_H37Rv | LAM | 0 | 0 | |||
| 14722_7#92_H37Rv | LAM | 0 | 0 | |||
| 14722_7#93_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14722_7#94_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_7#95_H37Rv | V483G, G594E | S450L | Other small clades | 1 | 1 | |
| 14722_8#1_H37Rv | T400N, S450L, V695L | T | 1 | 1 | ||
| 14722_8#10_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 14722_8#11_H37Rv | N698K | S450L | LAM | 1 | 1 | |
| 14722_8#12_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#13_H37Rv | V183G | S450L | Beijing | 1 | 1 | |
| 14722_8#14_H37Rv | I885V | S450L | LAM | 1 | 1 | |
| 14722_8#15_H37Rv | A1047P | S450L | LAM | 1 | 1 | |
| 14722_8#16_H37Rv | L516P, G594E | S450L | Haarlem | 1 | 1 | |
| 14722_8#17_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14722_8#18_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14722_8#19_H37Rv | G594E | D120G, H445Y | Haarlem | 1 | 1 | |
| 14722_8#2_H37Rv | R741S | H445D | LAM | 1 | 1 | |
| 14722_8#20_H37Rv | P1040L | S450L | LAM | 1 | 1 | |
| 14722_8#21_H37Rv | V483G | S450L | LAM | 1 | 1 | |
| 14722_8#22_H37Rv | N698K | S450L | LAM | 1 | 1 | |
| 14722_8#23_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_8#24_H37Rv | N698S | S450L | LAM | 1 | 1 | |
| 14722_8#25_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#26_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 14722_8#27_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_8#28_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#29_H37Rv | E207K, H445Y | LAM | 1 | 1 | ||
| 14722_8#3_H37Rv | G945V | S450L | LAM | 1 | 1 | |
| 14722_8#30_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#31_H37Rv | N698S | S450L | LAM | 1 | 1 | |
| 14722_8#32_H37Rv | T812I | S450L, A692T | LAM | 1 | 1 | |
| 14722_8#33_H37Rv | S450F | LAM | 1 | 1 | ||
| 14722_8#34_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#35_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_8#36_H37Rv | G594E | H445L | Haarlem | 1 | 1 | |
| 14722_8#37_H37Rv | G571R, G594E | S450L | Haarlem | 1 | 1 | |
| 14722_8#38_H37Rv | G594E, A734V | S450L | Haarlem | 1 | 1 | |
| 14722_8#39_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_8#4_H37Rv | T812I | A692T | LAM | 1 | 1 | |
| 14722_8#40_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 14722_8#41_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14722_8#42_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#43_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 14722_8#44_H37Rv | G594E | H445D | Haarlem | 1 | 1 | |
| 14722_8#45_H37Rv | G594E | S450L | Other small clades | 1 | 1 | |
| 14722_8#46_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#47_H37Rv | Q429H, D435Y | LAM | 1 | 1 | ||
| 14722_8#48_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_8#49_H37Rv | LAM | 0 | 1 | |||
| 14722_8#5_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#50_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14722_8#51_H37Rv | Other small clades | 0 | 1 | |||
| 14722_8#52_H37Rv | LAM | 0 | 1 | |||
| 14722_8#53_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14722_8#54_H37Rv | S450L, L731P | LAM | 1 | 1 | ||
| 14722_8#55_H37Rv | W484S | S450L | Beijing | 1 | 1 | |
| 14722_8#56_H37Rv | I491V | S450L, A692T | LAM | 1 | 1 | |
| 14722_8#57_H37Rv | V483G | S450L | LAM | 1 | 1 | |
| 14722_8#58_H37Rv | P1040R | S450L | LAM | 1 | 1 | |
| 14722_8#59_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14722_8#6_H37Rv | V1124E | S450L | LAM | 1 | 1 | |
| 14722_8#60_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#61_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#62_H37Rv | D435V, V695L | T | 1 | 1 | ||
| 14722_8#63_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 14722_8#64_H37Rv | D435V | Beijing | 1 | 1 | ||
| 14722_8#65_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#66_H37Rv | E49A | D435V | LAM | 1 | 1 | |
| 14722_8#67_H37Rv | LAM | 0 | 1 | |||
| 14722_8#68_H37Rv | H445C | LAM | 1 | 1 | ||
| 14722_8#69_H37Rv | N698S | S450L | LAM | 1 | 1 | |
| 14722_8#7_H37Rv | D435V | Beijing | 1 | 1 | ||
| 14722_8#70_H37Rv | V1252L | S450L, A692T | LAM | 1 | 1 | |
| 14722_8#71_H37Rv | G594E | Haarlem | 0 | 1 | ||
| 14722_8#72_H37Rv | G594E | Haarlem | 0 | 1 | ||
| 14722_8#73_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#74_H37Rv | S450L, K891E | T | 1 | 1 | ||
| 14722_8#75_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14722_8#76_H37Rv | G594E | D435V | Haarlem | 1 | 1 | |
| 14722_8#77_H37Rv | T812I | S450L, A692T | LAM | 1 | 1 | |
| 14722_8#78_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#79_H37Rv | G594E | D435V | Haarlem | 1 | 1 | |
| 14722_8#8_H37Rv | N698S | S450L | LAM | 1 | 1 | |
| 14722_8#80_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14722_8#81_H37Rv | P1040A | S450L, V695L | T | 1 | 1 | |
| 14722_8#82_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#83_H37Rv | D435V | LAM | 1 | 1 | ||
| 14722_8#84_H37Rv | K312E, S450L | LAM | 1 | 1 | ||
| 14722_8#85_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#86_H37Rv | V483G, G594E | S450L | Other small clades | 1 | 1 | |
| 14722_8#87_H37Rv | R741S | S450L | LAM | 1 | 1 | |
| 14722_8#88_H37Rv | G594E | S450F | Other small clades | 1 | 1 | |
| 14722_8#89_H37Rv | E207K, H445Y | LAM | 1 | 1 | ||
| 14722_8#9_H37Rv | T812I | S450L, A692T | LAM | 1 | 1 | |
| 14722_8#90_H37Rv | E757A | S450L, A692T | LAM | 1 | 1 | |
| 14722_8#91_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_8#92_H37Rv | Q409R, S450L | LAM | 1 | 1 | ||
| 14722_8#93_H37Rv | G594E, A734V | S450L | Haarlem | 1 | 1 | |
| 14722_8#94_H37Rv | S450L | LAM | 1 | 1 | ||
| 14722_8#95_H37Rv | G594E | Y308D, S450L | Other small clades | 1 | 1 | |
| 14892_2#1_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14892_2#10_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14892_2#11_H37Rv | L452P | LAM | 1 | 0 | ||
| 14892_2#12_H37Rv | D435V | Beijing | 1 | 1 | ||
| 14892_2#15_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14892_2#16_H37Rv | H445D | LAM | 1 | 1 | ||
| 14892_2#17_H37Rv | LAM | 0 | 0 | |||
| 14892_2#18_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14892_2#2_H37Rv | Beijing | 0 | 0 | |||
| 14892_2#20_H37Rv | T | 0 | 0 | |||
| 14892_2#21_H37Rv | LAM | 0 | 0 | |||
| 14892_2#22_H37Rv | Other small clades | 0 | 0 | |||
| 14892_2#23_H37Rv | LAM | 0 | 0 | |||
| 14892_2#25_H37Rv | R1163H | LAM | 0 | 0 | ||
| 14892_2#26_H37Rv | G594E | S450M | Haarlem | 1 | 0 | |
| 14892_2#27_H37Rv | G594E | S450M | Haarlem | 1 | 1 | |
| 14892_2#28_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#29_H37Rv | N698S | S450L | LAM | 1 | 1 | |
| 14892_2#30_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14892_2#31_H37Rv | LAM | 0 | 0 | |||
| 14892_2#32_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#33_H37Rv | LAM | 0 | 0 | |||
| 14892_2#34_H37Rv | P45L, S450L | LAM | 1 | 1 | ||
| 14892_2#35_H37Rv | V483A | Q432P | LAM | 1 | 1 | |
| 14892_2#36_H37Rv | G594E, G388A | E250G | Haarlem | 0 | 0 | |
| 14892_2#37_H37Rv | D435V | LAM | 1 | 0 | ||
| 14892_2#39_H37Rv | G388A, R741S | H445D | LAM | 1 | 1 | |
| 14892_2#4_H37Rv | LAM | 0 | 0 | |||
| 14892_2#40_H37Rv | LAM | 0 | 0 | |||
| 14892_2#41_H37Rv | G594E | D435V | Haarlem | 1 | 1 | |
| 14892_2#42_H37Rv | LAM | 0 | 0 | |||
| 14892_2#43_H37Rv | G388A, R741S | H445D | LAM | 1 | 1 | |
| 14892_2#44_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14892_2#45_H37Rv | I851V, I997V | S450L | LAM | 1 | 1 | |
| 14892_2#46_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14892_2#48_H37Rv | G594E | S450M | Haarlem | 1 | 0 | |
| 14892_2#49_H37Rv | D435Y V695L | T | 1 | 0 | ||
| 14892_2#5_H37Rv | H445Y | Beijing | 1 | 1 | ||
| 14892_2#50_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#51_H37Rv | S450L, E550G | EAM | 1 | 1 | ||
| 14892_2#52_H37Rv | EAM | 0 | 0 | |||
| 14892_2#53_H37Rv | EAM | 0 | 0 | |||
| 14892_2#54_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14892_2#57_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14892_2#58_H37Rv | I491V | S450L, A692T | LAM | 1 | 1 | |
| 14892_2#59_H37Rv | V483G | S450L, V695L | T | 1 | 1 | |
| 14892_2#60_H37Rv | LAM | 0 | 0 | |||
| 14892_2#61_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14892_2#63_H37Rv | W484S | S450L | Beijing | 1 | 1 | |
| 14892_2#64_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#65_H37Rv | G594E | S450F | Other small clades | 1 | 1 | |
| 14892_2#66_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14892_2#67_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14892_2#68_H37Rv | G594E | Other small clades | 0 | 1 | ||
| 14892_2#69_H37Rv | P615L | S450L | LAM | 1 | 1 | |
| 14892_2#7_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14892_2#70_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14892_2#71_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14892_2#72_H37Rv | Other small clades | 0 | 1 | |||
| 14892_2#73_H37Rv | G594E | Haarlem | 0 | 1 | ||
| 14892_2#74_H37Rv | G594E | P45S, S450L | Haarlem | 1 | 1 | |
| 14892_2#75_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#76_H37Rv | E1033K | F424L, 149IF | LAM | 0 | 1 | |
| 14892_2#77_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#78_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#79_H37Rv | H445Y | Beijing | 1 | 1 | ||
| 14892_2#8_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#80_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14892_2#81_H37Rv | S450L | LAM | 1 | 1 | ||
| 14892_2#82_H37Rv | G594E | S450L | Haarlem | 1 | 1 | |
| 14892_2#83_H37Rv | D435F | Beijing | 1 | 1 | ||
| 14892_2#84_H37Rv | E207K, H445Y | LAM | 1 | 1 | ||
| 14892_2#85_H37Rv | L516P, G594E | S450L | Haarlem | 1 | 1 | |
| 14892_2#86_H37Rv | D435V | LAM | 1 | 1 | ||
| 14892_2#88_H37Rv | V483G | S450L, V695L | T | 1 | 1 | |
| 14892_2#89_H37Rv | S428T, G594E | V170F, I488L | Other small clades | 0 | 1 | |
| 14892_2#9_H37Rv | R741S | S450F | LAM | 1 | 1 | |
| 14892_2#90_H37Rv | G594E, A734V | S450L | Haarlem | 1 | 1 | |
| 14892_2#91_H37Rv | V483G | S450L | LAM | 1 | 1 | |
| 14892_2#92_H37Rv | E207K, H445Y | LAM | 1 | 1 | ||
| 14892_2#93_H37Rv | S450L, V496M, V695L | T | 1 | 1 | ||
| 14892_2#94_H37Rv | G594E, A734V | S450L | Haarlem | 1 | 1 | |
| 14892_2#95_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#1_H37Rv | LAM | 0 | 0 | |||
| 14893_2#10_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#11_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#12_H37Rv | G594E | S450L | Haarlem | 1 | 1 | |
| 14893_2#13_H37Rv | LAM | 0 | 0 | |||
| 14893_2#14_H37Rv | G594E | Haarlem | 0 | 1 | ||
| 14893_2#15_H37Rv | V695L | T | 0 | 0 | ||
| 14893_2#16_H37Rv | Haarlem | 0 | 0 | |||
| 14893_2#17_H37Rv | G594E | L452P | Haarlem | 1 | 0 | |
| 14893_2#18_H37Rv | LAM | 0 | 1 | |||
| 14893_2#19_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#2_H37Rv | LAM | 0 | 0 | |||
| 14893_2#20_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14893_2#21_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 14893_2#22_H37Rv | G594E | I491F | Other small clades | 0 | 1 | |
| 14893_2#23_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14893_2#24_H37Rv | LAM | 0 | 0 | |||
| 14893_2#25_H37Rv | S450L | LAM | 1 | 1 | ||
| 14893_2#26_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#27_H37Rv | V695L | T | 0 | 0 | ||
| 14893_2#28_H37Rv | T | 0 | 1 | |||
| 14893_2#29_H37Rv | H445D, V695L | T | 1 | 1 | ||
| 14893_2#3_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#30_H37Rv | G594E | V170F | Other small clades | 0 | 1 | |
| 14893_2#31_H37Rv | I965V | Caprae | 0 | 0 | ||
| 14893_2#32_H37Rv | V695L | T | 0 | 0 | ||
| 14893_2#33_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#34_H37Rv | S450L, L731P | LAM | 1 | 1 | ||
| 14893_2#35_H37Rv | LAM | 0 | 0 | |||
| 14893_2#36_H37Rv | T812I | S450L, A692T | LAM | 1 | 1 | |
| 14893_2#37_H37Rv | N698S, R741S | S450L | LAM | 1 | 1 | |
| 14893_2#38_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14893_2#39_H37Rv | LAM | 0 | 0 | |||
| 14893_2#4_H37Rv | A286V, S450L | LAM | 1 | 1 | ||
| 14893_2#40_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14893_2#41_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#42_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#43_H37Rv | G594E, P1040A | S450L, R827C | Haarlem | 1 | 1 | |
| 14893_2#44_H37Rv | LAM | 0 | 0 | |||
| 14893_2#45_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14893_2#46_H37Rv | I491V | S450L, A692T | LAM | 1 | 1 | |
| 14893_2#47_H37Rv | G594E | S450L, G973D | Haarlem | 1 | 1 | |
| 14893_2#48_H37Rv | S450L, L731P | LAM | 1 | 1 | ||
| 14893_2#49_H37Rv | G594E | H445D | Haarlem | 1 | 1 | |
| 14893_2#5_H37Rv | A172V | EAI2-Manilla | 0 | 0 | ||
| 14893_2#50_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 14893_2#51_H37Rv | G594E | Other small clades | 0 | 0 | ||
| 14893_2#52_H37Rv | V483G | S450L, V695L | T | 1 | 1 | |
| 14893_2#53_H37Rv | LAM | 0 | 0 | |||
| 14893_2#54_H37Rv | A788V | D435F | Beijing | 1 | 1 | |
| 14893_2#55_H37Rv | D435F | Beijing | 1 | 1 | ||
| 14893_2#56_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#57_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#58_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 14893_2#59_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 14893_2#6_H37Rv | G594E, V1252L | S450L | Haarlem | 1 | 1 | |
| 14893_2#60_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#61_H37Rv | S450L, Q980R | LAM | 1 | 1 | ||
| 14893_2#62_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#63_H37Rv | G594E | D435V | Haarlem | 1 | 1 | |
| 14893_2#64_H37Rv | G594E | E250G | Haarlem | 0 | 1 | |
| 14893_2#65_H37Rv | LAM | 0 | 0 | |||
| 14893_2#66_H37Rv | S450L, I480V, A692T | LAM | 1 | 1 | ||
| 14893_2#67_H37Rv | G594E | Haarlem | 0 | 0 | ||
| 14893_2#68_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#69_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#7_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#70_H37Rv | L452P, V695L | T | 1 | 1 | ||
| 14893_2#71_H37Rv | G594E | S450L | Haarlem | 1 | 1 | |
| 14893_2#72_H37Rv | V483G | S450L, V695L | T | 1 | 1 | |
| 14893_2#73_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#74_H37Rv | G594E, V1206G, D1218A | S450W | Haarlem | 1 | 1 | |
| 14893_2#75_H37Rv | V1252M | S450L | LAM | 1 | 1 | |
| 14893_2#76_H37Rv | G594E | Haarlem | 0 | 1 | ||
| 14893_2#77_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#78_H37Rv | Beijing | 0 | 1 | |||
| 14893_2#79_H37Rv | V517L | S450L | LAM | 1 | 1 | |
| 14893_2#8_H37Rv | P1040A | S450L | LAM | 1 | 1 | |
| 14893_2#80_H37Rv | G332R | S450L | LAM | 1 | 1 | |
| 14893_2#81_H37Rv | H445Y | Beijing | 1 | 1 | ||
| 14893_2#82_H37Rv | E207K, H445Y | LAM | 1 | 1 | ||
| 14893_2#83_H37Rv | D435V | Beijing | 1 | 1 | ||
| 14893_2#84_H37Rv | G594E | D435V | Haarlem | 1 | 1 | |
| 14893_2#85_H37Rv | G594E | S450L | Other Small Clades | 1 | 1 | |
| 14893_2#86_H37Rv | W484S | S450L | Beijing | 1 | 1 | |
| 14893_2#87_H37Rv | V483A, G594E | S450L | Other small clades | 1 | 1 | |
| 14893_2#88_H37Rv | V483G | S450L, V695L | T | 1 | 1 | |
| 14893_2#89_H37Rv | W484S | S450L | Beijing | 1 | 1 | |
| 14893_2#9_H37Rv | L507V | S450L | T | 1 | 1 | |
| 14893_2#90_H37Rv | V483A | S450L | Beijing | 1 | 1 | |
| 14893_2#91_H37Rv | D435V | LAM | 1 | 1 | ||
| 14893_2#92_H37Rv | G594E | S428R, H445P | Other small clades | 1 | 1 | |
| 14893_2#93_H37Rv | V483A, G594E | S450L | Other small clades | 1 | 1 | |
| 14893_2#94_H37Rv | G571R, G594E | S450L | Haarlem | 1 | 1 | |
| 14893_2#95_H37Rv | V483G | S450L | LAM | 1 | 1 | |
| 14893_3#65_H37Rv | S450L, A692T | LAM | 1 | 1 | ||
| 14893_3#66_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 15277_3#50_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 15277_3#51_H37Rv | R741S, V1039A | S450L | NA | 0 | 0 | |
| 15277_3#52_H37Rv | S450L, V695L | T | 1 | 1 | ||
| 15277_3#53_H37Rv | S450L, E550G | LAM | 1 | 1 | ||
| 15277_3#55_H37Rv | V517L | S450L | NA | 0 | 0 | |
| 15277_3#57_H37Rv | S450L | LAM | 1 | 1 |
RIF: Rifampicin, RRDR: RIF resistance-determining region
Figure 1:
Representation of our 469-strain dataset [detailed in Table 1], classified according to resistance/susceptibility to rifampicin, presence/absence of mutations in rpoB, and presence/absence of mutations in rpoC or rpoA. The number in brackets indicates the number of strains with certain characteristics (i.e., there are 9 rifampicin-resistant strains that have a mutation in rpoC/rpoA but do not harbor mutations in rpoB)
175 (54%) strains that were RIF resistant and harbored mutations in rpoB had at least one mutation in rpoA or rpoC, suggesting that these may be compensatory mutations. 41 (41.8%) RIF-susceptible strains with no mutations in rpoB had at least one mutation in rpoC or rpoA. The presence of these 41 mutations may suggest the occurrence of random mutations, not necessarily associated with bacterial fitness.
Strikingly, 11 RIF-susceptible strains of different spoligotypes harbored rpoB mutations in the critical region (RRDR) [Table 1]. These mutations represent 3.4% of the 327 strains harboring a mutation in the RRDR. Five of these 11 strains also harbored the G594E mutation in rpoC [Table 2]. Notably, G594E appears only in the Haarlem genotype among these strains. This suggests that a very small percentage (3.4%) of mutations in the RRDR may not be associated with RIF resistance.
Table 2:
Mutations in the rpoB critical region (rifampicin resistance-determining region) not associated with rifampicin resistance
| Strain number | Mutation in rpoC |
Mutation in RRDR |
Spoligotyping |
|---|---|---|---|
| 14722_6#36_H37Rv | 0 | D435F | Beijing |
| 14722_6#41_H37Rv | 0 | D435F | Beijing |
| 14722_6#53_H37Rv | 0 | D435V | LAM |
| 14892_2#11_H37Rv | 0 | L452P | LAM |
| 14892_2#37_H37Rv | 0 | D435V | LAM |
| 14892_2#49_H37Rv | 0 | D435Y, V695L | T |
| 14722_6#17_H37Rv | G594E | L452P | Haarlem |
| 14722_7#75_H37Rv | G594E | L452P | Haarlem |
| 14892_2#26_H37Rv | G594E | S450M | Haarlem |
| 14892_2#48_H37Rv | G594E | S450M | Haarlem |
| 14893_2#17_H37Rv | G594E | L452P | Haarlem |
Trait present: Mutation in amino acid code, Trait absent: 0.
RIF: Rifampicin, RRDR: RIF resistance-determining region
Rifampicin susceptibility and genotyping
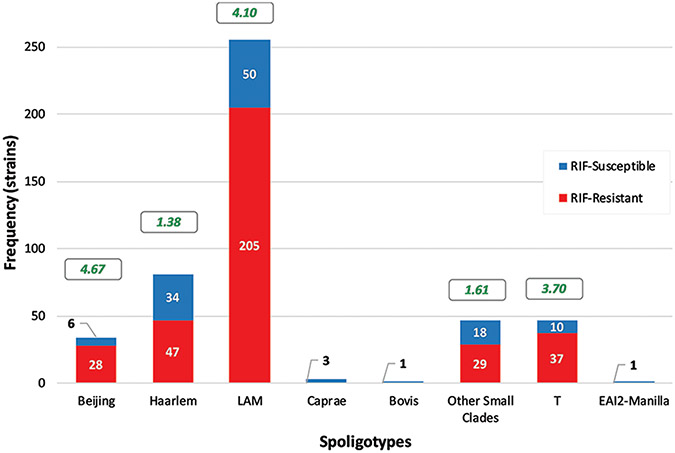
The distribution of RIF-resistant strains (i.e., the RIF-resistant/RIF-susceptible strains ratio) was similar (1.4–4.6) within the different spoligotype clades [Figure 2]. This suggests that our investigation is not biased toward a specific genotype. LAM was the predominant genotype (54.4%), followed by Haarlem (17.3%), other small clades and T (10.03%), Beijing (7.3%), Caprae (0.64%), and finally Bovis and EAI2-Manilla (0.21%).
Figure 2:
Frequency histogram of the 469 strains divided by spoligotype and sorted according to susceptibility to rifampicin. Red represents resistance while blue represents susceptibility. The white numbers represent the number of strains (frequency) that belong to certain spoligotypes and are susceptible/resistant to rifampicin. Black numbers have been added for contrast purposes but serve the same role. Green numbers in boxes represent the ratio of rifampicin resistant/rifampicin susceptible strains of each spoligotype
Mutations in rpoA and rpoC associated with rifampicin resistant strains
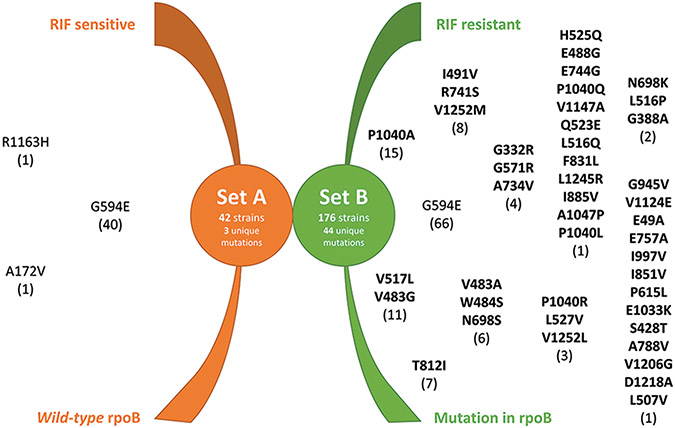
The first group generated for analysis [setA, Supplementary Table 1] comprised a total of 43 strains, while the second group [set B, Supplementary Table 2] comprised 176 strains. Both groups had a variety of rpoC mutations, as depicted in Figure 3.
Figure 3:
Graphical representation of rpoC mutations in Set A (mutations in rifampicin susceptible strains with wild-type rpoB, shown in orange on the left) and Set B (mutations in rifampicin-resistant strains harboring rpoB mutations, shown in green on the right). Mutations in bold are those found exclusively in Set B and thus possible compensatory mutations
Interestingly, G594E appeared in both groups, being related to strains associated with RIF resistance and mutations in rpoB, but also to strains with a wild-type rpoB and RIF susceptibility. Other studies also found this mutation and excluded it from their analyses.[14,16,17,20,22,23]
Forty-four mutations found exclusively in set B [bold in Figure 3] were considered to be potentially compensatory. Nine (22%) of these mutations have been previously described [Supplementary Table 3]: N698K,[17] I491V,[17] H525Q,[17] P1040R,[17] L516P,[16,19] V1252M,[17] V483A,[16,19] V483G,[16,17,19] and V1252 L.[17] The remaining 35 (78%) were novel.
Only three mutations in the rpoA gene were found (T187A, G31S, and V183G), all of which have been reported elsewhere.[16,17,19] Our literature search identified a total of 29 compensatory mutations in rpoA.
Distribution of the potentially compensatory mutations along the rpoC protein sequence
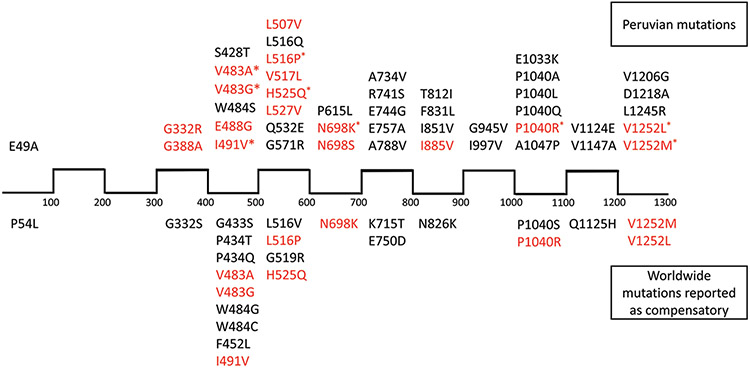
A high frequency of mutations occurred between amino acids 400–500, 500–600, and 1000–1100 (a total of 21 mutations) [Figure 4], suggesting the existence of hotspots. 47% of all the potentially compensatory mutations proposed in this study arose in these regions. E49A and P54 L were isolated from the rest, located within the first 100 amino acids of the protein. The distribution of the mutations found here and those reported previously was found to be remarkably similar (P = 0.2318, Kolmogorov–Smirnov nonparametric test).
Figure 4:
Linear distribution of the identified putative rpoC compensatory mutations throughout the protein sequence (scale: 100 amino acids per space). The upper part of the image lists the mutations found in our analysis, while the lower part shows mutations reported in the scientific literature. Mutations in red are those already reported elsewhere, but not necessarily underwent experimental confirmation of compensatory behavior. Upper mutations marked with an asterisk (*) are those already reported as compensatory by experimental assays
To better analyze this distribution, we classified the amino acids into the following groups: basic (R, K, and H), acidic (D and E), polar (G, S, Y, C, Q, T, and N), and nonpolar (F, L, W, P, I, M, V, and A). Based on this, we found that 54% of the potentially compensatory mutations found in rpoC preserve their physical–chemical properties despite the amino acid change.
Distribution of the potential compensatory rpoC mutations on the three-dimensional structure of Mycobacterium tuberculosis RNApol
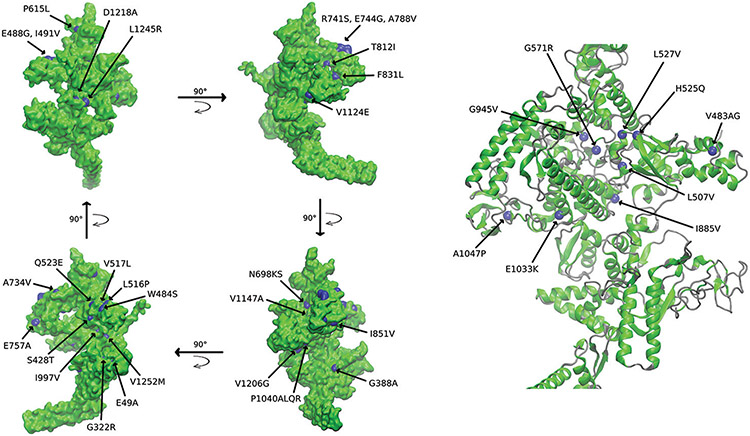
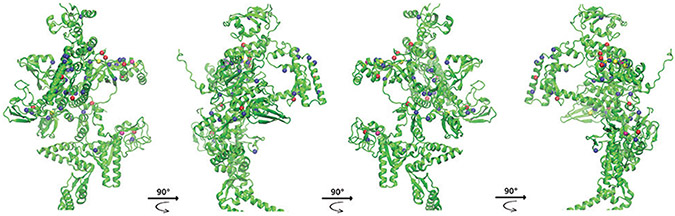
The potentially compensatory mutations that we described above [Supplementary Table 3] were mainly distributed on the surface of the three-dimensional (3D)-crystal structure of the β’ subunit (82%), and 9 were buried below the surface [Figure 5]. Only one of the buried mutated sites, H525Q, has been previously reported to be compensatory.[17] E49A, which was visibly separated from the rest in the primary structure of rpoC, can be seen on the side of the subunit 3D-structure, distant from the main cluster of mutations [Figure 5].
Figure 5:
Crystal of the Mycobacterium tuberculosis β’ subunit (rpoC), represented according to its surface and its secondary structure. The surface, β sheets, and α helices are shown in green, while loops are colored gray. The mutations found in our study are either pictured in blue patches (surface) or as Van der Waals spheres (alpha carbon of buried residues only, radius scale = 1.4 Å). The arrows in the left panel indicate the direction of the rotation of the protein along the Y axis
β’ RNApol has a domain that resembles the shape of an “arm” that hosts a particularly large number of mutations. The “arm” is best visible on the right side of the structure [Figure 6]. Our data show that some of the putative compensatory mutations fall in similar spots as those that have been previously reported, in particular those located in this “arm”. Interestingly, the reported mutation P54 L appears hidden below the surface along with E49A [Figure 6]. We also found that two of the proposed mutations, G332R and W484S, occur in regions where other mutations have already been reported.
Figure 6:
Crystal of the Mycobacterium tuberculosis β’ subunit (rpoC), represented by its secondary structure. The β sheets and α helices are shown in green, while loops are colored gray. Different types of mutations are shown in different colors, as follows: Blue for the putative compensatory mutations found in our study, red for previously reported compensatory mutations, yellow for mutations reported in the literature and also found in our study, and magenta for mutations found in this study that had another amino acid change (for the same position) reported in the literature. All the mutations are represented as Van der Waals spheres (alpha carbon only, radius scale = 1.4 Å). The arrows indicate the direction of the rotation of the protein along the Y axis
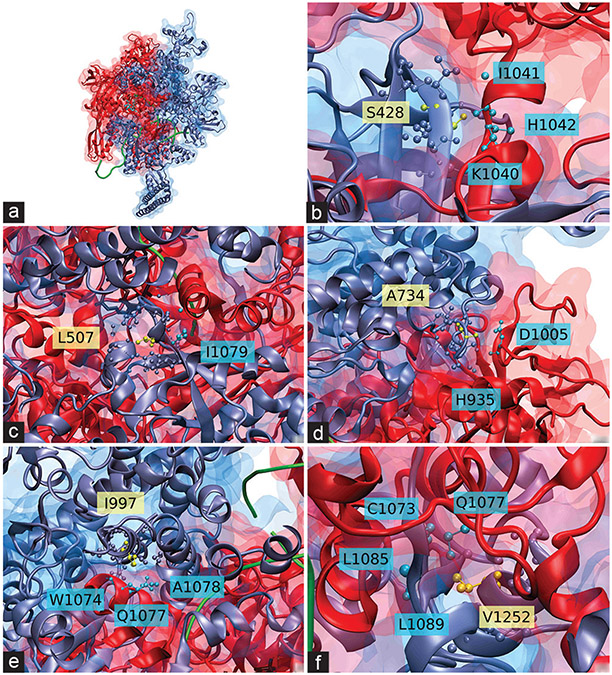
The 3D-crystal structure of β’ interacting with the β subunit confirms that several mutations fall in the regions of interaction (S428T, L507V, A734V, I997V, and V1252 LM) [Figure 7]. These represent 13.5% of the surface mutations. All of the putative compensatory mutations that we identified in the rpoC/rpoB contact surface conserved their physical–chemical properties during the amino acid change [Figure 7].
Figure 7:
Crystal structure of Mycobacterium tuberculosis transcription complex showing interaction of β’ (rpoC) and β (rpoB) subunits. Proteins are represented by their secondary structure and their surface; rpoC is shown in iceblue and rpoB is shown in red. Two DNA strands are shown in green as a ribbon, along with a rifampin molecule in light blue (panel a) or black (panels b– f). rpoC residues whose sites would carry compensatory mutations are marked in yellow in Corey-Pauling-Koltun model (CPK) representation (side chain included). rpoB residues within 5.00 Å (6.00 Å for panel c) of the marked rpoC residues are shown in cyan in CPK representation. rpoC residues within that distance are shown in iceblue in CPK representation, (a) Provides a full, general view of the interaction between the subunits, while the rest of the panels show the putative rpoC compensatory mutations interacting with close residues in rpoB: S428T (b), L507V (c), A734V (d), I997V (e), V1252 LM (f). The amino acids are indicated according to standard code, according to the subunit they belong (yellow for rpoC, blue for rpoB)
Discussion
This study describes 35 novel mutations in rpoC, associated with RIF resistance and rpoB mutations, detected in Peruvian clinical isolates of M. tuberculosis. These mutations may serve to compensate for any loss of fitness caused by a mutation in rpoB, as reported in other studies.[8,10,13-15] Of note, some were found to be associated with controversial mutations in rpoB that have been identified in RIF-susceptible MTB strains.
The similarity in the distribution of the potentially compensatory rpoC mutations found in Peru compared to those reported elsewhere is striking. The spatial distribution along the protein sequence reveals a similar, almost mirrored pattern, suggesting that compensatory mutations in rpoC preferentially occur in certain positions. These cluster-like regions could be related to functional aspects of the protein or may be structurally critical. According to the Pfam protein domain family database,[30-32] these high-frequency mutation zones correspond to specific domains. The first zone (amino acids 400–500, second domain) contains the protein’s active site, the second zone (amino acids 500–600, third domain) contains the pore thought to act as a channel for nucleotides, and the third zone (amino acids 1000–1100, fifth domain) includes the discontinuous cleft required to form the channel where DNA is bound. Mutations occurring in these critical regions may cause significant changes in function.
Surface mutations are more likely to interact with other subunits at a quaternary structure level or with other proteins. Furthermore, a surface mutation would not significantly affect the tertiary structure of the protein, whereas a deep mutation may destabilize the whole subunit. Thus, it would be favorable for compensatory mutations to be located on the surface of the protein, even more on the interaction sites. In our study, the position of the novel mutations on the crystal structure revealed that 82.2% fall on the surface of the β’ subunit.
Moreover, only six mutations were found in the region of interaction between the surfaces of β and β’, which constitute our most likely candidates for compensation, as a mutation at this site would produce a conformational change sufficient to affect the binding between the subunits but not significant enough to disrupt the inner core. Two of these six mutations have already been reported, and although W484S was not found in the literature, W484G/C has in fact been previously described. Moreover, as noted before and according to our expectations that mutations would result in minimal (but significant) structural changes, every mutation that we reported as a candidate conserves the physical–chemical properties of the original amino acid, favoring the maintenance of the protein’s stability.[33] Several mutations found in this study are very close to mutations reported elsewhere, and in some cases, even at the same position. This may also indicate the preference of certain locations for the emergence of compensatory mutations.
Only 20.5% of the mutations that we identified coincided with previously reported compensatory mutations.[16,17,19] This is likely to be due to the large genotypic diversity among Peruvian M. tuberculosis strains. Of note, in our study, most of the isolates (250) corresponded to the “LAM” spoligotype.
We identified just three mutations in rpoA, which is thought to be, along with rpoC, the most important site of RIF-resistant compensatory mutations. All three have been previously reported. This is in keeping with previous findings that the majority of compensatory mutations arise in rpoC,[16,17,19] suggesting that these may be more effective at recovering loss of fitness arising from mutations in rpoB.[16,21-23]
Furthermore, we found mutations in rpoC that were not associated with any mutations in rpoB but were present in RIF-resistant strains. It is likely that the appearance of these mutations is random, but further studies are necessary to clarify their true nature.
We reported 11 RIF-susceptible strains harboring mutations in the RRDR, with 5 harboring rpoC G594E [Table 2]. This is unlikely to be due to incorrect DST results, as MODS is a highly sensitive (98%) and specific (98.6%) method.[26] Rather, this finding led us to further analyze our data in search of patterns in the distribution of rpoC G594E. We found it to be present in 17.3% of RIF-resistant and 39.8% of RIF-susceptible strains, being by far the most common mutation. G594E has been described in strains with no rpoB mutations and is a SNP present in all strains from a group within the Haarlem genotype, which would mean that it arises independently of rpoB mutations and RIF resistance. Taken together, these features make it unlikely that G594E is associated with drug resistance, as broadly stated before.[14,16,17,20,22,23]
In view of this, we found G594E in both of our analysis groups: set A (rpoC mutations in RIF-susceptible strains without rpoB mutations) and set B (rpoC mutations in RIF-resistant strains harboring rpoB mutations). Furthermore, we found it in the five previously mentioned RIF-susceptible strains with mutations in the RRDR [Table 2]. Three of these harbor rpoB L452P, described elsewhere.[19,34] The remaining two harbor rpoB S450M, which is similar to the widely-known S450 L[10,17-19,34] and their described variants S450F/Q/Y/W.[19,21,34-38] The detection of rpoB S450M is puzzling because a Ser to Met mutation would require changes in two nucleotides (TCG → ATG), making it unlikely to occur spontaneously. It is possible that S450F occurred first (TCG → TTG), followed by a change from TTG → ATG, resulting in the end of a Ser to Met change. It is notable that both amino acids are nonpolar; that is, they have similar physicochemical properties. We could not find any other study describing S450M, so we encourage further investigation of the role and evolution of this mutation, as our theory of its development is mere speculation.
Nonetheless, we cannot avoid discussing the dichotomy regarding the appearance of rpoC G594E. Considering all of the above, we propose another possibility toward its appearance, which is that under certain conditions some mutations in rpoB do not cause RIF resistance but may still decrease the fitness of the bacteria. This would subdue the bacteria to selective pressure, leading to the positive selection of a compensatory mutation in rpoC. Until now, most investigations have attributed the unlikeliness of G594E being associated with drug resistance to its uncharacteristic appearance, but believe that there may be other ways for this to happen and thus we cannot draw any conclusions about the true nature of this mutation until more is known.
Our study identifies mutations that may have a compensatory role, but there is a need for experimental confirmation of the effect of each mutation as some of these are likely to arise at random. Previous studies have used methods such as the measurement of the catalytic activity and rate of bacterial growth.[14-18] We suggest that the use of site-directed mutagenesis techniques, such as the emerging CRISPR-Cas9 technology,[39] would be useful to assess the fitness of mutant strains.
Conclusion
This study identifies 35 putative novel compensatory mutations in the β’ subunit of M. tuberculosis RNApol. Six of these (S428T, L507V, A734V, I997V, and V1252 LM) are considered most likely to have a compensatory role, as they fall in the interaction zone of the two subunits and the mutation did not lead to any change in the protein’s physical–chemical properties. Further studies assessing the fitness change in strains harboring these mutations are needed to confirm their potential compensatory role. In addition, it would be interesting to evaluate the evolutionary relationship of the various SNPs found in this study.
Supplementary Material
Financial support and sponsorship
This research was funded by the Wellcome Trust (Ref: 099805/Z/12/Z), the Grand Challenge Canada (GCC Number 0687-01-10), and the LOREAL-UNESCO-CONCYTEC 2014 award.
PS was supported by a Wellcome Trust Intermediate fellowship.
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.World Health Organization. World Health Statistics 2016: Monitoring Health for the SDGs, Sustainable Development Goals. Geneva: World Health Organization. Available from: https://apps.who.int/iris/bitstream/handle/10665/324835/9789241565707-eng.pdf7ua=1. [Last updated on 2020 Jan 13; Last accessed on 2019 Jan 20]. [Google Scholar]
- 2.Silva C, Bermúdez V, Arraiz N, Bermúdez F, Rodríguez M, Valdelamar L, et al. Fármacos de primera línea utilizados en el tratamiento de la tuberculosis. Arch Venez Farmacol Y Ter 2007;26:21–6. [Google Scholar]
- 3.World Health Organization. Global Tuberculosis Report. Geneva: World Health Organization; Available from: https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf?ua=1. [Last updated on 2020 Jan 13; Last accessed on 2019 Jan 20]. [Google Scholar]
- 4.World Health Organization. Tuberculosis. Available from: https://www.who.int/en/news-room/fact-sheets/detail/tuberculosis. [Last updated on 2020 Jan 13; Last accessed on 2019 Jan 20]
- 5.World Health Organization. Drug resistant TB-Data by WHO Region. Available from: http://apps.who.int/gho/data/view.main.MDRTBWHOREG?lang=en. [Last updated on 2020 Jan 13; Last accessed on 2019 Jan 20]
- 6.Ministry of Health. Lima: Analysis of the Epidemiological Situation in Peru. Available from: http://bvs.minsa.gob.pe/local/MINSA/3446.pdf. [Last updated on 2020 Jan 13; Last accessed on 2019 Jan 20] [Google Scholar]
- 7.Mendoza L, Rodríguez H, Rodríguez E, Tarazona C. Conocimientos básicos sobre antibacterianos. 1st ed. Maracaibo: Editorial Astro Data; 1999. p. 73–5. [Google Scholar]
- 8.Koch A, Mizrahi V, Warner DF. The impact of drug resistance on Mycobacterium tuberculosis physiology: What can we learn from rifampicin? Emerg Microbes Infect 2014;3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes D, Brandis G. Rifampicin resistance: Fitness costs and the significance of compensatory evolution. Antibiotics (Basel) 2013;2:206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alifano P, Palumbo C, Pasanisi D, Talà A. Rifampicin-resistance, rpoB polymorphism and RNA polymerase genetic engineering. J Biotechnol 2015;202:60–77. [DOI] [PubMed] [Google Scholar]
- 11.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 1993;341:647–50. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein BP. Resistance to rifampicin: A review. J Antibiot (Tokyo) 2014;67:625–30. [DOI] [PubMed] [Google Scholar]
- 13.Brandis G, Hughes D. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J Antimicrob Chemother 2013;68:2493–7. [DOI] [PubMed] [Google Scholar]
- 14.Song T, Park Y, Shamputa IC, Seo S, Lee SY, Jeon HS, et al. Fitness costs of rifampicin resistance in Mycobacterium tuberculosis are amplified under conditions of nutrient starvation and compensated by mutation in the β’ subunit of RNA polymerase. Mol Microbiol 2014;91:1106–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2004;48:1289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 2011;44:106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen KA, Abeel T, Manson McGuire A, Desjardins CA, Munsamy V, Shea TP, et al. Evolution of extensively drug-resistant tuberculosis over four decades: Whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from kwazulu-natal. PLoS Med 2015;12:e1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandis G, Wrande M, Liljas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol 2012;85:142–51. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet 2013;45:1255–60. [DOI] [PubMed] [Google Scholar]
- 20.Ali A, Hasan Z, McNerney R, Mallard K, Hill-Cawthorne G, Coll F, et al. Whole genome sequencing based characterization of extensively drug-resistant Mycobacterium tuberculosis isolates from Pakistan. PLoS One 2015;10:e0117771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res 2012;22:735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vos M, Müller B, Borrell S, Black PA, van Helden PD, Warren RM, et al. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother 2013;57:827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li QJ, Jiao WW, Yin QQ, Xu F, Li JQ, Sun L, et al. Compensatory mutations of rifampin resistance are associated with transmission of multidrug-resistant Mycobacterium tuberculosis Beijing genotype strains in China. Antimicrob Agents Chemother 2016;60:2807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandjean L, Gilman RH, Iwamoto T, Köser CU, Coronel J, Zimic M, et al. Convergent evolution and topologically disruptive polymorphisms among multidrug-resistant tuberculosis in Peru. PLoS One 2017;12:e0189838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caviedes L, Lee TS, Gilman RH, Sheen P, Spellman E, Lee EH, et al. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. The Tuberculosis Working Group in Peru. J Clin Microbiol 2000;38:1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minion J, Leung E, Menzies D, Pai M. Microscopic-observation drug susceptibility and thin layer agar assays for the detection of drug resistant tuberculosis: A systematic review and meta-analysis. Lancet Infect Dis 2010;10:688–98. [DOI] [PubMed] [Google Scholar]
- 27.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 1997;35:907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin W, Mandal S, Degen D, Liu Y, Ebright YW, Li S, et al. Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition. Mol Cell 2017;66:169–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph 1996;14:33–8, 27-8. [DOI] [PubMed] [Google Scholar]
- 30.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res 2016;44:D279–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, Darst SA, et al. Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the beta and beta’ subunits of Escherichia coli RNA polymerase. J Biol Chem 1996;271:27969–74. [DOI] [PubMed] [Google Scholar]
- 32.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 2001;292:1863–76. [DOI] [PubMed] [Google Scholar]
- 33.Ladunga I, Smith RF. Amino acid substitutions preserve protein folding by conserving steric and hydrophobicity properties. Protein Eng. Des. Sel 1997;10:187–96. [DOI] [PubMed] [Google Scholar]
- 34.Casali N, Broda A, Harris SR, Parkhill J, Brown T, Drobniewski F. Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: A Retrospective Observational Study. PLoS Med 2016;13:e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salamon H, Yamaguchi KD, Cirillo DM, Miotto P, Schito M, Posey J, et al. Integration of published information into a resistance-associated mutation database for Mycobacterium tuberculosis. J Infect Dis 2015;211 Suppl 2:S50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang C, Luo T, Shen X, Wu J, Gan M, Xu P, et al. Transmission of multidrug-resistant Mycobacterium tuberculosis in Shanghai, China: A retrospective observational study using whole-genome sequencing and epidemiological investigation. Lancet Infect Dis 2017;17:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ullah I, Ahmad W, Shah AA, Shahzada A, Tahir Z, Qazi O, et al. Detection of rifampicin resistance of Mycobacterium tuberculosis using multiplex allele specific polymerase chain reaction (MAS-PCR) in Pakistan. Infect Genet Evol 2019;71:42–6. [DOI] [PubMed] [Google Scholar]
- 38.Miotto P, Cabibbe AM, Borroni E, Degano M, Cirillo DM. Role of Disputed Mutations in the rpoB Gene in Interpretation of Automated Liquid MGIT Culture Results for Rifampin Susceptibility Testing of Mycobacterium tuberculosis. J Clin Microbiol 2018;56:e01599–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol 2017;2:16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.