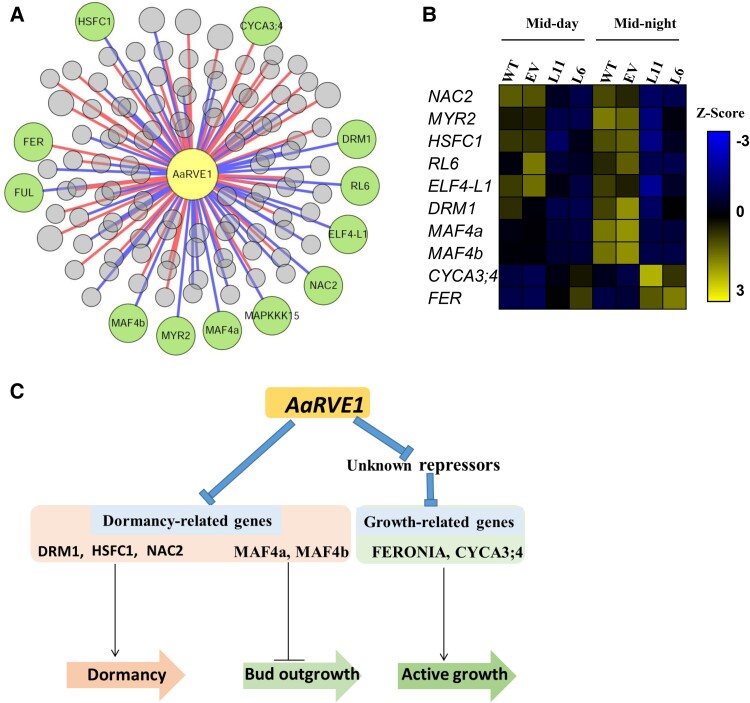
Figure 4.
AaRVE1 regulated multiple dormancy-related genes and cell division- and cell elongation-related genes. A, Co-expression network analysis of AaRVE1. The significantly DEGs identified by RNA-seq were subjected to co-expression analysis with the expression of the AaRVE1 gene in the transgenic plants L6 and L11, EV, and wild-types (WT) (Supplemental Table 2). Green nodes represent the dormancy-related genes and bud break-related genes. Red and blue edges represent positive and negative co-expression relationships, respectively (P < 0.05 and |Correlation coefficient| > 0.7). B, Expression profiles of the dormancy-related genes and bud break-related genes at mid-day and mid-night. The gene information can be found in Supplemental Table 2. C, Diagram showing the regulation of bud dormancy and bud break in the transgenic plants expressing AaRVE1. DRM1: DORMANCY-ASSOCIATED PROTEIN 1; NAC2: NO APICAL MERISTEM FAMILY PROTEIN; HSFC1: HEAT SHOCK TRANSCRIPTION FACTOR C1; MAF: MADS AFFECTING FLOWERING (MAF)-like; CYCA3; 4: CYCLIN A3; 4.