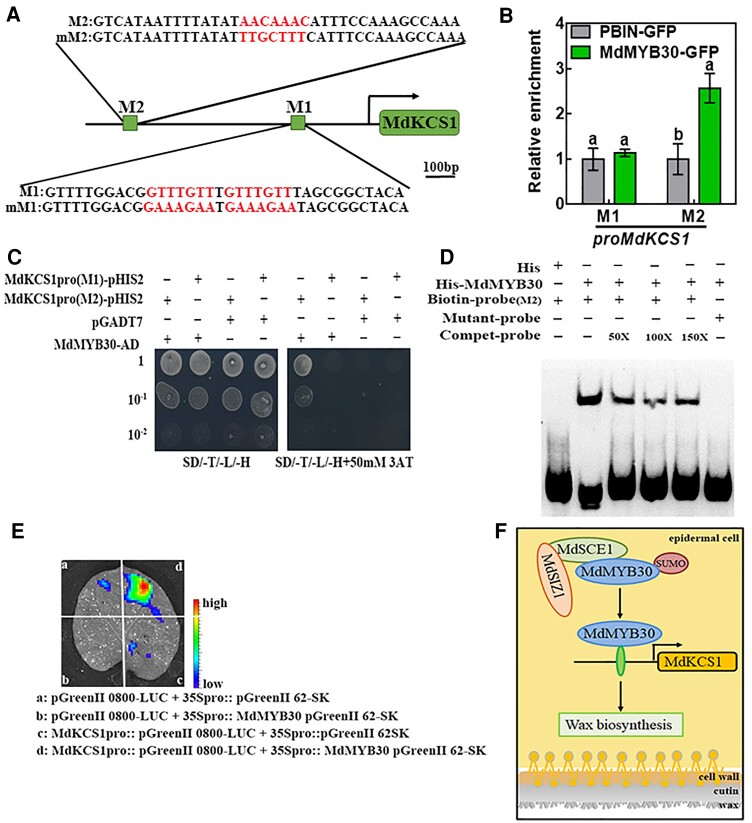
Figure 9.
MdMYB30 directly binds to the MdKCS1 promoter. A, A schematic diagram depicting the M1 and M2 motifs in the MdKCS1 promoter. Red letters indicate binding motifs and their corresponding mutation sequences. Scale bar, 100 bp. B, ChIP-qPCR analysis for testing the binding of MdMYB30 to the M1 and M2 motifs in the promoter sequence of MdKCS1. Enrichment of PBIN-GFP was used as a reference and set to 1. Data are mean ± Sd of three independent replicates. Different letters indicate significant differences (ANOVA, Student's t test; P < 0.05). C, Yeast one-hybrid assay to assess the binding of MdMYB30 to MdKCS1 promoter DNA. The yeast strains were grown on SD/-Trp/-Leu/-His/and SD/-Leu/-Trp/-His/ + 50 mM 3-AT media for 3 d. Empty pGADT7 vector was used as a negative control. The experiments were repeated three times, and similar results were obtained across all replicates. D, EMSA for testing the binding of MdMYB30 to the putative M2 motif in the MdKCS1 promoters. Competitive probe (Compet-probe) refers to the unlabeled probe. 50×, 100×, and 150× represent the multiples of the competitor probe. The AACAAAC (M2) motifs were mutated to TTGCTTT in M2. E, Transient expression assays showing that MdMYB30 activated the expression of MdKCS1pro::LUC. Representative images of Nicotiana benthamiana leaves 72 h after infiltration. F, Schematic diagram of the working model for the roles of MdSIZ1 and MdMYB30. MdSIZ1 positively regulates the level of MdMYB30 by promoting its SUMOylation. MdMYB30 directly binds to and activates the expression of MdKCS1, which positively regulates wax biosynthesis.