TABLE 2.
Ligands name, 3D structures, SMILE format and the virtual screening outputs.
| S. No. | Ligand | 3D structure | SMILE format | Binding score (kcal/mol) | H-bond residues |
|---|---|---|---|---|---|
| 1 | Procyanidin B2 |
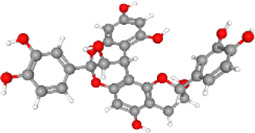
|
C1C(C(OC2=C1C(=CC3 =C2C4C(C(O3)(OC5= CC(=CC(=C45)O)O)C6=CC(=C(C=C6)O)O)O)O)C7=CC(=C(C=C7)O)O)O | −9.3 | Ala100, Arg107, Asn143, Gly145, and Arg146 |
| 2 | Luteolin-7-O-rutinoside |
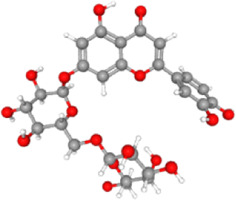
|
C1=CC(=C(C=C1C2=CC(=O)C3=C(C=C(C=C3O2)OC4C(C(C(C(O4)COC5C(C(C(C(O5)CO)O)O)O)O)O)O)O)O)O | −9.1 | Asp111, Asn143 and Arg146 |
