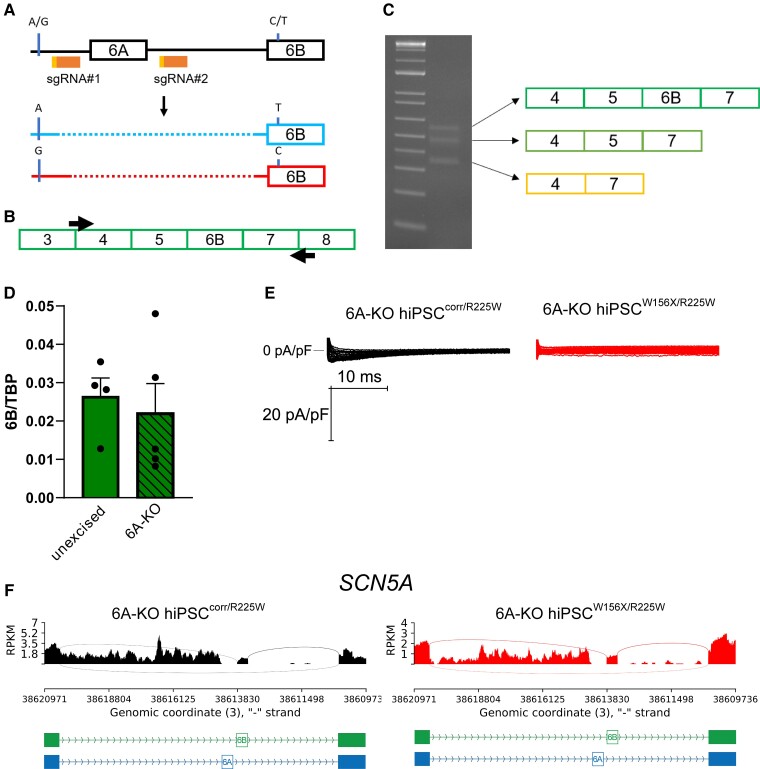
Figure 3.
Genetic excision of exon 6A does not increase adult SCN5A isoform expression. (A) Schematic representation of the strategy to excise SCN5A exon 6A using CRISPR/Cas9 with two sgRNAs (sgRNA#1 and #2, orange). In red and light blue, the results of excision (dotted line) in the hiPSCsW156X/R225W: the maternal allele (top, blue, with the mutated T in exon 6B and the A SNP in the intron between exon 5 and exon 6A) carried a 15 bp larger deletion upstream of exon 6A compared with the paternal allele (bottom, red, WT C in exon 6B and G SNP in 5-6A intron). (B) Schematic showing the primers (black arrows) used to amplify and sequencing cDNA from 6A-KO hiPSCcorr/R225W-CMs. (C) Gel electrophoresis of PCR products from (B) showing three bands corresponding to three transcript species as indicated by the arrows. (D) Bar graph of mean expression of exon 6B in unexcised and 6A-KO hiPSC-CMs. Data were normalized to TBP. n = 4. Dots: single values. (E) Representative INa traces recorded from 6A-KO hiPSCcorr/R225W and 6A-KO hiPSCW156X/R225W CMs showing an almost complete absence of the current. (F) Sashimi plots of RNA-seq data from 6A-KO hiPSCcorr/R225W and 6A-KO hiPSCW156X/R225W CMs showing the absence of exon 6A transcription but extensive transcription of the intronic region between exon 5 and exon 6B.