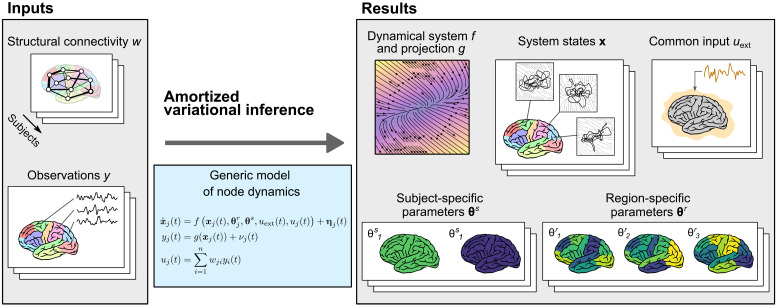
Fig. 1. Conceptual overview of the method.
The method allows us to perform a parameter inference for network models of brain dynamics, where the dynamical model of every node (or brain region) is initially unknown. As input (left), it expects structural connectivity matrices w for cohort of multiple subjects and corresponding observations of brain activity y (such as parcellated resting-state fMRI). Constrained by the structure of a generic model of regional dynamics (middle), it learns the dynamical model of node dynamics f and the state-to-observation projection model g. The dynamical model f is shared for all subjects and regions, but it depends on subject-specific parameters θs and region-specific parameters θr. These, too, are inferred from the data, together with the hidden states of the system x and the subject-specific external input uext, shared by all regions in a given subject. All of the system states, subject- and region-specific parameters, and the external input are inferred probabilistically as normal distributions, that is, we infer the mean and variance for each parameter.